Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
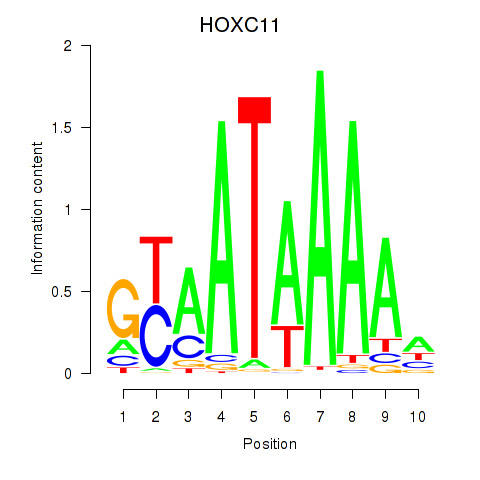
Results for HOXC11
Z-value: 0.60
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | HOXC11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC11 | hg19_v2_chr12_+_54366894_54366922 | -0.33 | 2.1e-01 | Click! |
Activity profile of HOXC11 motif
Sorted Z-values of HOXC11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_46679185 | 1.94 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 1.92 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr2_+_228678550 | 1.80 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chrX_-_105282712 | 1.46 |
ENST00000372563.1 |
SERPINA7 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chrX_+_107683096 | 1.27 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr5_-_111091948 | 1.17 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr16_-_67978016 | 1.10 |
ENST00000264005.5 |
LCAT |
lecithin-cholesterol acyltransferase |
| chr7_-_47579188 | 1.06 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr7_+_134464414 | 0.97 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr6_+_12290586 | 0.83 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr8_+_26435359 | 0.81 |
ENST00000311151.5 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr1_+_229440129 | 0.78 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr10_+_114709999 | 0.78 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_-_39684550 | 0.77 |
ENST00000455635.1 ENST00000361566.3 |
KRT19 |
keratin 19 |
| chr3_-_79816965 | 0.77 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr2_-_166930131 | 0.73 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr1_-_12677714 | 0.73 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr14_-_25479811 | 0.73 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr10_-_13523073 | 0.69 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr17_-_77924627 | 0.68 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr16_+_15031300 | 0.66 |
ENST00000328085.6 |
NPIPA1 |
nuclear pore complex interacting protein family, member A1 |
| chr10_+_114710516 | 0.63 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr16_+_16472912 | 0.57 |
ENST00000530217.2 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr1_+_167298281 | 0.57 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr16_+_14805546 | 0.56 |
ENST00000552140.1 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr19_+_1440838 | 0.55 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr19_+_49468558 | 0.53 |
ENST00000331825.6 |
FTL |
ferritin, light polypeptide |
| chr7_+_134464376 | 0.53 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr16_+_16429787 | 0.52 |
ENST00000331436.4 ENST00000541593.1 |
AC138969.4 |
Protein PKD1P1 |
| chr16_-_21431078 | 0.48 |
ENST00000458643.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr16_-_18466642 | 0.47 |
ENST00000541810.1 ENST00000531453.2 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr7_-_27169801 | 0.46 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr2_+_211421262 | 0.44 |
ENST00000233072.5 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr22_+_39916558 | 0.42 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr1_+_183774240 | 0.37 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr19_-_10420459 | 0.36 |
ENST00000403352.1 ENST00000403903.3 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
| chr8_-_18666360 | 0.35 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chrX_-_107682702 | 0.35 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr12_-_24102576 | 0.32 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr8_-_57123815 | 0.30 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr1_+_52682052 | 0.27 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr7_-_11871815 | 0.27 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr2_-_207023918 | 0.26 |
ENST00000455934.2 ENST00000449699.1 ENST00000454195.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_+_173116225 | 0.25 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr1_-_237167718 | 0.25 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr13_+_97928395 | 0.24 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr2_+_166428839 | 0.22 |
ENST00000342316.4 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr12_-_123201337 | 0.21 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr7_-_27205136 | 0.21 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr1_+_87794150 | 0.19 |
ENST00000370544.5 |
LMO4 |
LIM domain only 4 |
| chr1_+_209859510 | 0.18 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr1_+_26485511 | 0.16 |
ENST00000374268.3 |
FAM110D |
family with sequence similarity 110, member D |
| chr6_+_108487245 | 0.16 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr6_+_15246501 | 0.15 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chrM_+_4431 | 0.15 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_64029879 | 0.15 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr8_+_104831554 | 0.15 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr10_+_26505179 | 0.14 |
ENST00000376261.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr9_-_124990680 | 0.14 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr6_-_41703952 | 0.14 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr9_-_124989804 | 0.14 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr3_+_89156799 | 0.14 |
ENST00000452448.2 ENST00000494014.1 |
EPHA3 |
EPH receptor A3 |
| chr2_-_51259292 | 0.14 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chr5_-_34043310 | 0.14 |
ENST00000231338.7 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
| chr4_-_74088800 | 0.13 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr12_+_1800179 | 0.13 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr13_-_30424821 | 0.13 |
ENST00000380680.4 |
UBL3 |
ubiquitin-like 3 |
| chr10_+_115614370 | 0.13 |
ENST00000369301.3 |
NHLRC2 |
NHL repeat containing 2 |
| chr17_-_46806540 | 0.12 |
ENST00000290295.7 |
HOXB13 |
homeobox B13 |
| chr7_+_22766766 | 0.12 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr11_-_118305921 | 0.12 |
ENST00000532619.1 |
RP11-770J1.4 |
RP11-770J1.4 |
| chr13_-_46626847 | 0.11 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr22_+_41697520 | 0.11 |
ENST00000352645.4 |
ZC3H7B |
zinc finger CCCH-type containing 7B |
| chr2_-_51259229 | 0.11 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chr2_+_109204909 | 0.11 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr17_+_7461580 | 0.10 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_+_47894023 | 0.10 |
ENST00000358474.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr1_-_7913089 | 0.10 |
ENST00000361696.5 |
UTS2 |
urotensin 2 |
| chr2_-_50574856 | 0.10 |
ENST00000342183.5 |
NRXN1 |
neurexin 1 |
| chr13_-_28545276 | 0.09 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chr1_+_23345930 | 0.09 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr8_+_76452097 | 0.09 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr4_+_160188889 | 0.09 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_63606373 | 0.08 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr8_-_20040638 | 0.08 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr1_+_62439037 | 0.08 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr4_-_107957454 | 0.07 |
ENST00000285311.3 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
| chr17_-_39646116 | 0.07 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chrX_+_99899180 | 0.07 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr13_+_109248500 | 0.07 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chrX_-_10851762 | 0.06 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr1_+_23345943 | 0.06 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr6_+_31683117 | 0.06 |
ENST00000375825.3 ENST00000375824.1 |
LY6G6D |
lymphocyte antigen 6 complex, locus G6D |
| chr7_+_108210012 | 0.06 |
ENST00000249356.3 |
DNAJB9 |
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr3_-_45838011 | 0.06 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr7_+_141811539 | 0.06 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr2_-_209010874 | 0.06 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr2_+_177015122 | 0.05 |
ENST00000468418.3 |
HOXD3 |
homeobox D3 |
| chr3_-_45837959 | 0.05 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr14_+_56127989 | 0.05 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr2_-_40739501 | 0.05 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_-_81399329 | 0.05 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr5_+_145718587 | 0.04 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chr8_-_20040601 | 0.04 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr9_+_105757590 | 0.04 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr8_+_134203273 | 0.03 |
ENST00000250160.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr2_+_27440229 | 0.03 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr11_-_27723158 | 0.03 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr1_+_209878182 | 0.03 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr7_-_151217166 | 0.03 |
ENST00000496004.1 |
RHEB |
Ras homolog enriched in brain |
| chr17_+_7533439 | 0.03 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr2_+_176987088 | 0.02 |
ENST00000249499.6 |
HOXD9 |
homeobox D9 |
| chr3_+_89156674 | 0.02 |
ENST00000336596.2 |
EPHA3 |
EPH receptor A3 |
| chr16_-_2314222 | 0.01 |
ENST00000566397.1 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
| chr20_+_4702548 | 0.01 |
ENST00000305817.2 |
PRND |
prion protein 2 (dublet) |
| chr8_-_59412717 | 0.01 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr17_+_7461613 | 0.00 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_136678123 | 0.00 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr2_+_210444142 | 0.00 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr8_+_134203303 | 0.00 |
ENST00000519433.1 ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 1.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.5 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.7 | GO:0043194 | voltage-gated sodium channel complex(GO:0001518) axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.6 | 1.8 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 0.8 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 1.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.3 | 0.8 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 1.4 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.4 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.3 | GO:0021853 | cerebral cortex tangential migration(GO:0021800) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.8 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 1.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 0.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.5 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.4 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |