Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
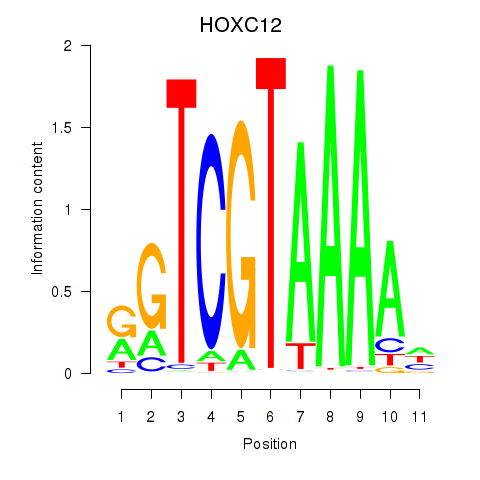
Results for HOXC12_HOXD12
Z-value: 1.23
Transcription factors associated with HOXC12_HOXD12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC12
|
ENSG00000123407.3 | HOXC12 |
|
HOXD12
|
ENSG00000170178.5 | HOXD12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD12 | hg19_v2_chr2_+_176964458_176964540 | -0.26 | 3.3e-01 | Click! |
Activity profile of HOXC12_HOXD12 motif
Sorted Z-values of HOXC12_HOXD12 motif
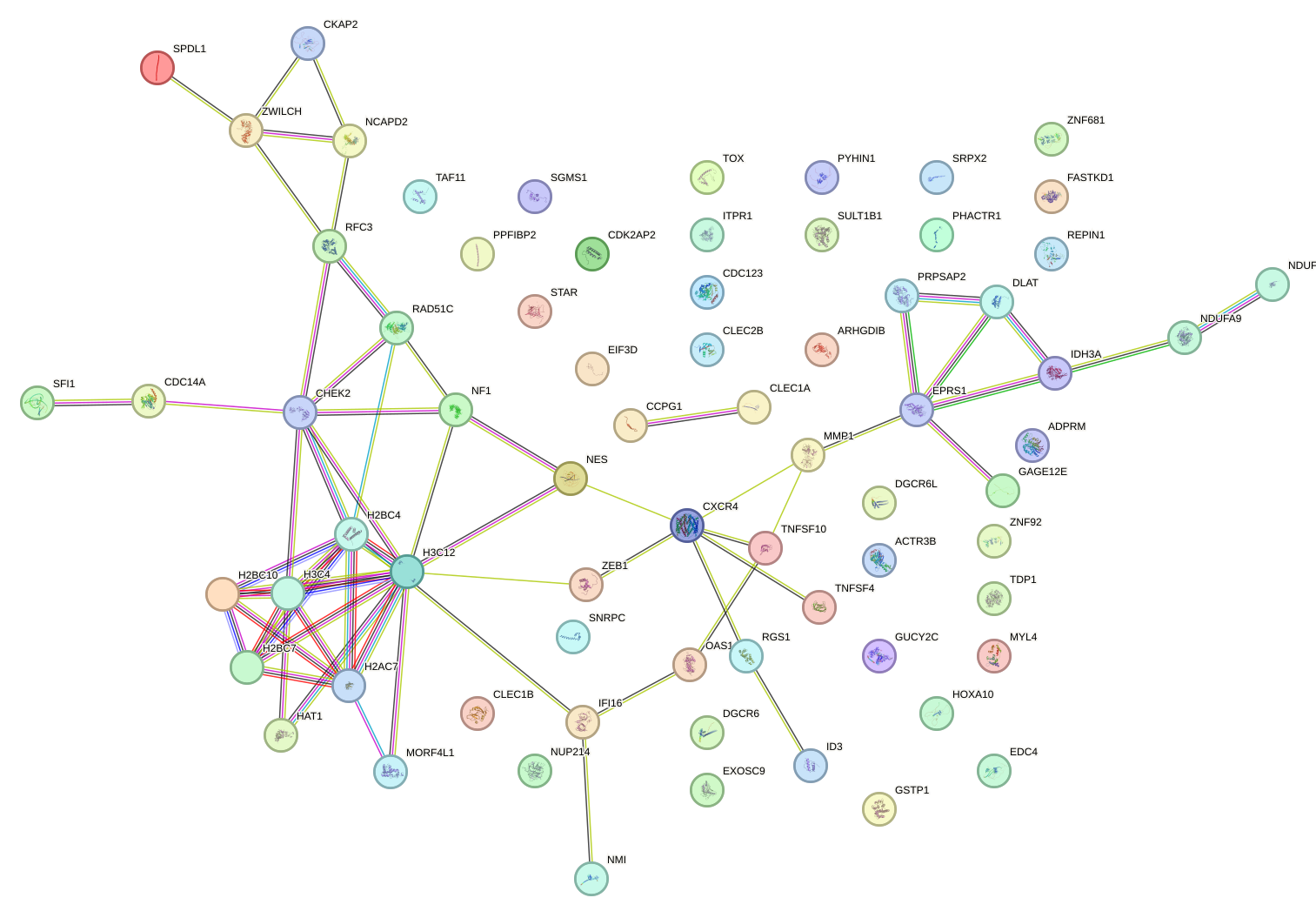
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC12_HOXD12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_15918618 | 2.23 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr7_+_150065278 | 1.55 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr6_+_13272904 | 1.46 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr18_-_53253323 | 1.45 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr1_-_246670519 | 1.45 |
ENST00000388985.4 ENST00000490107.1 |
SMYD3 |
SET and MYND domain containing 3 |
| chr2_-_152146385 | 1.43 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr17_+_45286706 | 1.41 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr18_-_53253112 | 1.40 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chrX_+_49294472 | 1.30 |
ENST00000361446.5 |
GAGE12B |
G antigen 12B |
| chr10_-_52383644 | 1.30 |
ENST00000361781.2 |
SGMS1 |
sphingomyelin synthase 1 |
| chr12_-_15114492 | 1.27 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr10_+_31608054 | 1.26 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr12_-_15114603 | 1.23 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_+_111896320 | 1.18 |
ENST00000531306.1 ENST00000537636.1 |
DLAT |
dihydrolipoamide S-acetyltransferase |
| chr7_+_152456904 | 1.11 |
ENST00000537264.1 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
| chr6_+_26199737 | 1.10 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr1_+_158975744 | 1.06 |
ENST00000426592.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_+_67351019 | 1.04 |
ENST00000398606.3 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr1_+_192544857 | 1.03 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr22_-_20307532 | 1.02 |
ENST00000405465.3 ENST00000248879.3 |
DGCR6L |
DiGeorge syndrome critical region gene 6-like |
| chr19_-_48752812 | 0.99 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr19_-_48753104 | 0.98 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr22_+_23134974 | 0.94 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr1_-_220219775 | 0.89 |
ENST00000609181.1 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr12_-_10007448 | 0.89 |
ENST00000538152.1 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr1_+_158900568 | 0.89 |
ENST00000458222.1 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr8_-_33370607 | 0.88 |
ENST00000360742.5 ENST00000523305.1 |
TTI2 |
TELO2 interacting protein 2 |
| chr22_+_23101182 | 0.87 |
ENST00000390312.2 |
IGLV2-14 |
immunoglobulin lambda variable 2-14 |
| chr19_-_23941680 | 0.86 |
ENST00000402377.3 |
ZNF681 |
zinc finger protein 681 |
| chr12_+_113354341 | 0.85 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_+_169010638 | 0.83 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr19_+_21265028 | 0.81 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
| chr2_+_172778952 | 0.81 |
ENST00000392584.1 ENST00000264108.4 |
HAT1 |
histone acetyltransferase 1 |
| chr8_-_38008783 | 0.75 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr11_+_67351213 | 0.74 |
ENST00000398603.1 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr2_-_136875712 | 0.74 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr3_-_172241250 | 0.73 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr7_+_64838712 | 0.73 |
ENST00000328747.7 ENST00000431504.1 ENST00000357512.2 |
ZNF92 |
zinc finger protein 92 |
| chr6_-_26199499 | 0.73 |
ENST00000377831.5 |
HIST1H3D |
histone cluster 1, H3d |
| chr15_+_79166065 | 0.72 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr7_+_64838786 | 0.70 |
ENST00000450302.2 |
ZNF92 |
zinc finger protein 92 |
| chr11_+_111896090 | 0.70 |
ENST00000393051.1 |
DLAT |
dihydrolipoamide S-acetyltransferase |
| chr1_-_173176452 | 0.70 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_+_152456829 | 0.69 |
ENST00000377776.3 ENST00000256001.8 ENST00000397282.2 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
| chr1_-_220220000 | 0.66 |
ENST00000366923.3 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr9_+_134065506 | 0.62 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr1_-_23886285 | 0.60 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr3_+_4535025 | 0.58 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr9_+_2029019 | 0.58 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_+_90422239 | 0.58 |
ENST00000393452.3 ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1 |
tyrosyl-DNA phosphodiesterase 1 |
| chr2_-_136873735 | 0.55 |
ENST00000409817.1 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chrX_+_99899180 | 0.55 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr11_-_107729887 | 0.54 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr11_-_5255861 | 0.54 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr2_-_225811747 | 0.52 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr12_+_4758264 | 0.51 |
ENST00000266544.5 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr6_+_34725263 | 0.51 |
ENST00000374018.1 ENST00000374017.3 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr12_+_6603253 | 0.50 |
ENST00000382457.4 ENST00000545962.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr17_+_56769924 | 0.50 |
ENST00000461271.1 ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C |
RAD51 paralog C |
| chr22_-_36925186 | 0.50 |
ENST00000541106.1 ENST00000455547.1 ENST00000432675.1 |
EIF3D |
eukaryotic translation initiation factor 3, subunit D |
| chr7_+_90339169 | 0.49 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr5_-_176433693 | 0.48 |
ENST00000507513.1 ENST00000511320.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr12_+_120105558 | 0.48 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr5_-_176433582 | 0.48 |
ENST00000506128.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr2_-_170430366 | 0.48 |
ENST00000453153.2 ENST00000445210.1 |
FASTKD1 |
FAST kinase domains 1 |
| chr22_-_36924944 | 0.48 |
ENST00000405442.1 ENST00000402116.1 |
EIF3D |
eukaryotic translation initiation factor 3, subunit D |
| chr15_+_66797627 | 0.47 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr3_+_179322573 | 0.46 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_-_169680745 | 0.46 |
ENST00000236147.4 |
SELL |
selectin L |
| chr4_-_70626314 | 0.46 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr11_-_67275542 | 0.45 |
ENST00000531506.1 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr6_-_26199471 | 0.45 |
ENST00000341023.1 |
HIST1H2AD |
histone cluster 1, H2ad |
| chr22_-_32058166 | 0.45 |
ENST00000435900.1 ENST00000336566.4 |
PISD |
phosphatidylserine decarboxylase |
| chr18_-_5544241 | 0.44 |
ENST00000341928.2 ENST00000540638.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr6_+_34725181 | 0.43 |
ENST00000244520.5 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr17_+_29664830 | 0.43 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr15_+_66797455 | 0.43 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr7_-_27219849 | 0.42 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr12_-_10251603 | 0.42 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr17_+_18759612 | 0.42 |
ENST00000432893.2 ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2 |
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr11_-_102668879 | 0.42 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr6_-_34855773 | 0.40 |
ENST00000420584.2 ENST00000361288.4 |
TAF11 |
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr22_+_18893736 | 0.40 |
ENST00000331444.6 |
DGCR6 |
DiGeorge syndrome critical region gene 6 |
| chr4_+_122722466 | 0.40 |
ENST00000243498.5 ENST00000379663.3 ENST00000509800.1 |
EXOSC9 |
exosome component 9 |
| chr1_-_156647189 | 0.38 |
ENST00000368223.3 |
NES |
nestin |
| chr8_-_60031762 | 0.38 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr15_-_55657428 | 0.37 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr15_+_78441663 | 0.37 |
ENST00000299518.2 ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr11_+_7618413 | 0.37 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr17_-_8113542 | 0.36 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr16_+_67906919 | 0.36 |
ENST00000358933.5 |
EDC4 |
enhancer of mRNA decapping 4 |
| chr14_-_22005343 | 0.36 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr13_+_34392185 | 0.35 |
ENST00000380071.3 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr7_+_134331550 | 0.35 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr13_+_53030107 | 0.34 |
ENST00000490903.1 ENST00000480747.1 |
CKAP2 |
cytoskeleton associated protein 2 |
| chr22_-_29107919 | 0.33 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr10_+_12238171 | 0.32 |
ENST00000378900.2 ENST00000442050.1 |
CDC123 |
cell division cycle 123 |
| chr17_+_46184911 | 0.32 |
ENST00000580219.1 ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11 |
sorting nexin 11 |
| chr4_+_39460620 | 0.32 |
ENST00000340169.2 ENST00000261434.3 |
LIAS |
lipoic acid synthetase |
| chr1_+_100818009 | 0.30 |
ENST00000370125.2 ENST00000361544.6 ENST00000370124.3 |
CDC14A |
cell division cycle 14A |
| chr17_+_10600894 | 0.30 |
ENST00000379774.4 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr11_-_3400442 | 0.30 |
ENST00000429541.2 ENST00000532539.1 |
ZNF195 |
zinc finger protein 195 |
| chr8_+_27950619 | 0.30 |
ENST00000542181.1 ENST00000524103.1 ENST00000537665.1 ENST00000380353.4 ENST00000520288.1 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
| chr10_+_12237924 | 0.29 |
ENST00000429258.2 ENST00000281141.4 |
CDC123 |
cell division cycle 123 |
| chr12_+_10658201 | 0.29 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr4_+_106631966 | 0.29 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr12_+_67663056 | 0.28 |
ENST00000545606.1 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
| chr6_-_112080256 | 0.28 |
ENST00000462856.2 ENST00000229471.4 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr4_-_76957214 | 0.28 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr19_-_22018966 | 0.28 |
ENST00000599906.1 ENST00000354959.4 |
ZNF43 |
zinc finger protein 43 |
| chr1_+_199996702 | 0.27 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr8_+_1993152 | 0.27 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr20_-_44540686 | 0.26 |
ENST00000477313.1 ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP |
phospholipid transfer protein |
| chr16_+_20818020 | 0.26 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr17_-_34890732 | 0.26 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr3_+_179322481 | 0.26 |
ENST00000259037.3 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr19_+_58281014 | 0.26 |
ENST00000391702.3 ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586 |
zinc finger protein 586 |
| chr10_-_98031265 | 0.26 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr13_+_111855414 | 0.26 |
ENST00000375737.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr19_+_49496705 | 0.26 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr11_-_3400330 | 0.25 |
ENST00000427810.2 ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195 |
zinc finger protein 195 |
| chr10_+_51371390 | 0.25 |
ENST00000478381.1 ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B |
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr1_+_171750776 | 0.25 |
ENST00000458517.1 ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13 |
methyltransferase like 13 |
| chr19_+_21579908 | 0.25 |
ENST00000596302.1 ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493 |
zinc finger protein 493 |
| chr3_-_56502375 | 0.25 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr18_+_9103957 | 0.25 |
ENST00000400033.1 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr12_+_32832134 | 0.25 |
ENST00000452533.2 |
DNM1L |
dynamin 1-like |
| chr7_+_138145076 | 0.24 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr17_-_34890709 | 0.24 |
ENST00000544606.1 |
MYO19 |
myosin XIX |
| chr1_-_202896310 | 0.24 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr16_-_3450963 | 0.24 |
ENST00000573327.1 ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32 |
zinc finger and SCAN domain containing 32 |
| chr8_+_1993173 | 0.24 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr2_-_70475730 | 0.23 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_+_2443202 | 0.23 |
ENST00000258711.6 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr19_+_2785458 | 0.23 |
ENST00000307741.6 ENST00000585338.1 |
THOP1 |
thimet oligopeptidase 1 |
| chr10_-_75226166 | 0.23 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_159656437 | 0.23 |
ENST00000402432.3 |
FABP6 |
fatty acid binding protein 6, ileal |
| chr12_-_30848914 | 0.23 |
ENST00000256079.4 |
IPO8 |
importin 8 |
| chr11_-_32457075 | 0.23 |
ENST00000448076.3 |
WT1 |
Wilms tumor 1 |
| chr19_+_13885252 | 0.22 |
ENST00000221576.4 |
C19orf53 |
chromosome 19 open reading frame 53 |
| chr12_-_108154925 | 0.22 |
ENST00000228437.5 |
PRDM4 |
PR domain containing 4 |
| chr1_-_158656488 | 0.22 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr2_-_89521942 | 0.22 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr4_-_69536346 | 0.22 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chrX_-_15619076 | 0.22 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr9_-_77502636 | 0.22 |
ENST00000449912.2 |
TRPM6 |
transient receptor potential cation channel, subfamily M, member 6 |
| chr2_+_37571845 | 0.22 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr12_-_71182695 | 0.21 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr4_-_46911248 | 0.21 |
ENST00000355591.3 ENST00000505102.1 |
COX7B2 |
cytochrome c oxidase subunit VIIb2 |
| chr2_+_37571717 | 0.21 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr14_-_82089405 | 0.21 |
ENST00000554211.1 |
RP11-799P8.1 |
RP11-799P8.1 |
| chr2_-_88285309 | 0.21 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr10_-_98031310 | 0.20 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr2_-_86564776 | 0.20 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr1_-_53686261 | 0.20 |
ENST00000294360.4 |
C1orf123 |
chromosome 1 open reading frame 123 |
| chr10_-_99052382 | 0.20 |
ENST00000453547.2 ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1 ARHGAP19 |
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr4_-_46911223 | 0.20 |
ENST00000396533.1 |
COX7B2 |
cytochrome c oxidase subunit VIIb2 |
| chr10_-_51623203 | 0.19 |
ENST00000444743.1 ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23 |
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr7_+_138818490 | 0.19 |
ENST00000430935.1 ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26 |
tetratricopeptide repeat domain 26 |
| chr19_+_49496782 | 0.19 |
ENST00000601968.1 ENST00000596837.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr19_+_49497121 | 0.19 |
ENST00000413176.2 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr7_-_32529973 | 0.19 |
ENST00000410044.1 ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_-_156569754 | 0.19 |
ENST00000420343.1 |
MED7 |
mediator complex subunit 7 |
| chr13_-_79233314 | 0.18 |
ENST00000282003.6 |
RNF219 |
ring finger protein 219 |
| chr20_-_17539456 | 0.18 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chrX_-_133792480 | 0.18 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chrX_-_138724677 | 0.18 |
ENST00000370573.4 ENST00000338585.6 ENST00000370576.4 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr10_-_27443294 | 0.18 |
ENST00000396296.3 ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1 |
YME1-like 1 ATPase |
| chr6_+_57182400 | 0.18 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr8_+_107738240 | 0.18 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr3_-_186288097 | 0.18 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr18_+_72167096 | 0.18 |
ENST00000324301.8 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr5_-_146781153 | 0.17 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr14_+_88471468 | 0.17 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr1_+_12977513 | 0.17 |
ENST00000330881.5 |
PRAMEF7 |
PRAME family member 7 |
| chr4_-_120243545 | 0.17 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr5_-_175815565 | 0.17 |
ENST00000509257.1 ENST00000507413.1 ENST00000510123.1 |
NOP16 |
NOP16 nucleolar protein |
| chr4_-_69434245 | 0.17 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr1_-_35450897 | 0.17 |
ENST00000373337.3 |
ZMYM6NB |
ZMYM6 neighbor |
| chr3_-_17783990 | 0.17 |
ENST00000429383.4 ENST00000446863.1 ENST00000414349.1 ENST00000428355.1 ENST00000425944.1 ENST00000445294.1 ENST00000444471.1 ENST00000415814.2 |
TBC1D5 |
TBC1 domain family, member 5 |
| chr6_+_19837592 | 0.17 |
ENST00000378700.3 |
ID4 |
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr6_-_27782548 | 0.17 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr7_+_138145145 | 0.17 |
ENST00000415680.2 |
TRIM24 |
tripartite motif containing 24 |
| chr17_+_71228740 | 0.17 |
ENST00000268942.8 ENST00000359042.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr8_-_93029865 | 0.17 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_101491319 | 0.16 |
ENST00000342173.7 ENST00000488176.1 ENST00000370109.3 |
DPH5 |
diphthamide biosynthesis 5 |
| chr2_-_225362533 | 0.16 |
ENST00000451538.1 |
CUL3 |
cullin 3 |
| chr19_-_45996465 | 0.16 |
ENST00000430715.2 |
RTN2 |
reticulon 2 |
| chrY_+_26997726 | 0.16 |
ENST00000382296.2 |
DAZ4 |
deleted in azoospermia 4 |
| chr8_-_80942061 | 0.16 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr1_+_200993071 | 0.16 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr2_+_169926047 | 0.16 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr8_-_63998590 | 0.15 |
ENST00000260116.4 |
TTPA |
tocopherol (alpha) transfer protein |
| chr20_+_36932521 | 0.15 |
ENST00000262865.4 |
BPI |
bactericidal/permeability-increasing protein |
| chr9_+_133569108 | 0.15 |
ENST00000372358.5 ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2 |
exosome component 2 |
| chr2_+_170335924 | 0.14 |
ENST00000554017.1 ENST00000392663.2 ENST00000513963.1 |
BBS5 RP11-724O16.1 |
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr3_+_137906154 | 0.14 |
ENST00000466749.1 ENST00000358441.2 ENST00000489213.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr19_-_49955050 | 0.14 |
ENST00000262265.5 |
PIH1D1 |
PIH1 domain containing 1 |
| chr7_+_28452130 | 0.14 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr10_+_124913930 | 0.14 |
ENST00000368858.5 |
BUB3 |
BUB3 mitotic checkpoint protein |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 2.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.5 | 1.4 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.4 | 1.8 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.3 | 1.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 | 0.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.2 | 0.7 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 2.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.2 | 1.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 1.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 0.6 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 1.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 1.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.4 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.1 | 0.3 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 1.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.5 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.2 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 2.5 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 1.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 1.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.5 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.7 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 1.1 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.4 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 1.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.4 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 1.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 3.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.5 | 1.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 1.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 0.6 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 2.0 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.6 | 1.9 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.4 | 1.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.8 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.4 | 1.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.3 | 0.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 2.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |