Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
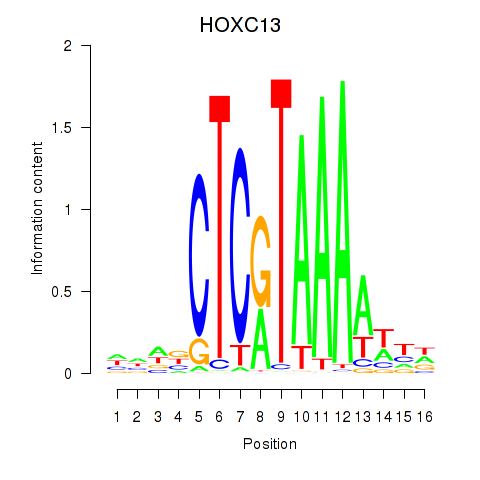
Results for HOXC13
Z-value: 0.99
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | HOXC13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.11 | 7.0e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_33041378 | 3.40 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr1_+_158979680 | 2.36 |
ENST00000368131.4 ENST00000340979.6 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 2.32 |
ENST00000368132.3 ENST00000295809.7 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_174128639 | 1.99 |
ENST00000251507.4 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_+_192544857 | 1.79 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr10_+_70847852 | 1.77 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr4_-_71532339 | 1.77 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_174844645 | 1.68 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr18_+_61554932 | 1.65 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr3_+_159557637 | 1.62 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr22_+_23101182 | 1.59 |
ENST00000390312.2 |
IGLV2-14 |
immunoglobulin lambda variable 2-14 |
| chr8_-_95274536 | 1.56 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr9_-_88715044 | 1.52 |
ENST00000388711.3 ENST00000466178.1 |
GOLM1 |
golgi membrane protein 1 |
| chr12_+_75874580 | 1.48 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr14_-_106963409 | 1.43 |
ENST00000390621.2 |
IGHV1-45 |
immunoglobulin heavy variable 1-45 |
| chr1_+_158815588 | 1.38 |
ENST00000438394.1 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr15_+_71185148 | 1.29 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr16_+_12059050 | 1.26 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr1_-_169680745 | 1.23 |
ENST00000236147.4 |
SELL |
selectin L |
| chr15_+_71228826 | 1.19 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr22_+_38142235 | 1.15 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr12_-_91539918 | 1.12 |
ENST00000548218.1 |
DCN |
decorin |
| chr16_+_33605231 | 1.11 |
ENST00000570121.2 |
IGHV3OR16-12 |
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr14_-_106668095 | 1.11 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr9_-_88714421 | 1.10 |
ENST00000388712.3 |
GOLM1 |
golgi membrane protein 1 |
| chr2_+_33359687 | 1.10 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359646 | 1.09 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr15_+_71184931 | 1.09 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr6_-_32784687 | 1.07 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr13_-_46964177 | 1.01 |
ENST00000389908.3 |
KIAA0226L |
KIAA0226-like |
| chrX_+_13707235 | 1.01 |
ENST00000464506.1 |
RAB9A |
RAB9A, member RAS oncogene family |
| chr18_-_52989217 | 0.98 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr18_-_52989525 | 0.97 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_-_225811747 | 0.96 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr5_+_150020214 | 0.93 |
ENST00000307662.4 |
SYNPO |
synaptopodin |
| chr10_-_79398250 | 0.90 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_112080256 | 0.87 |
ENST00000462856.2 ENST00000229471.4 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr1_+_43766642 | 0.85 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr1_-_89591749 | 0.82 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chr12_-_91546926 | 0.81 |
ENST00000550758.1 |
DCN |
decorin |
| chr12_-_4647606 | 0.80 |
ENST00000261250.3 ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4 |
chromosome 12 open reading frame 4 |
| chr7_-_41742697 | 0.77 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr10_+_13628933 | 0.76 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr14_-_106725723 | 0.73 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr12_-_92539614 | 0.68 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr10_-_6019984 | 0.67 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr7_+_80275953 | 0.64 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr5_-_146781153 | 0.64 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr18_-_5544241 | 0.64 |
ENST00000341928.2 ENST00000540638.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr21_+_37442239 | 0.60 |
ENST00000530908.1 ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1 |
carbonyl reductase 1 |
| chrX_-_135338503 | 0.60 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chr9_-_5304432 | 0.58 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr9_-_37034028 | 0.57 |
ENST00000520281.1 ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5 |
paired box 5 |
| chr4_-_120243545 | 0.55 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr7_+_80275752 | 0.52 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr8_-_93029865 | 0.52 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_+_102953608 | 0.50 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr19_+_13858593 | 0.46 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chrX_+_119737806 | 0.46 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chr1_+_84630645 | 0.45 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_-_46922659 | 0.44 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chr2_+_210444142 | 0.42 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr3_-_172241250 | 0.41 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr19_-_45982076 | 0.41 |
ENST00000423698.2 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr1_+_153003671 | 0.41 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr19_-_11456935 | 0.40 |
ENST00000590788.1 ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205 RAB3D |
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr19_-_54619006 | 0.39 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr9_-_95244781 | 0.39 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr15_+_79166065 | 0.38 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr21_-_27423339 | 0.38 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr10_-_79397740 | 0.37 |
ENST00000372440.1 ENST00000480683.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79398127 | 0.37 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_118797475 | 0.37 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr9_-_2844058 | 0.36 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr19_-_54618650 | 0.35 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr6_+_29426230 | 0.35 |
ENST00000442615.1 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr7_+_80275621 | 0.34 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr4_-_69536346 | 0.32 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr3_-_123339418 | 0.31 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr1_-_67266939 | 0.31 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr7_-_76255444 | 0.31 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr10_-_70287172 | 0.31 |
ENST00000539557.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr3_-_46037299 | 0.30 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr11_-_102576537 | 0.30 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr3_-_98241760 | 0.29 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chr12_+_67663056 | 0.27 |
ENST00000545606.1 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
| chr14_-_106816253 | 0.27 |
ENST00000390615.2 |
IGHV3-33 |
immunoglobulin heavy variable 3-33 |
| chr3_+_101498269 | 0.26 |
ENST00000491511.2 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr3_-_123339343 | 0.26 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr19_-_18632861 | 0.25 |
ENST00000262809.4 |
ELL |
elongation factor RNA polymerase II |
| chr10_-_70287231 | 0.24 |
ENST00000609923.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr5_-_79287060 | 0.23 |
ENST00000512560.1 ENST00000509852.1 ENST00000512528.1 |
MTX3 |
metaxin 3 |
| chr22_-_18111499 | 0.23 |
ENST00000413576.1 ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr22_-_32808194 | 0.21 |
ENST00000451746.2 ENST00000216038.5 |
RTCB |
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr12_-_52828147 | 0.20 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr5_-_180076580 | 0.20 |
ENST00000502649.1 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr3_+_129159039 | 0.20 |
ENST00000507564.1 ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr2_-_86850949 | 0.20 |
ENST00000237455.4 |
RNF103 |
ring finger protein 103 |
| chr15_-_78538025 | 0.20 |
ENST00000560817.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr11_+_7618413 | 0.20 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_+_101170563 | 0.19 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr17_-_8286484 | 0.19 |
ENST00000582556.1 ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26 |
ribosomal protein L26 |
| chr1_-_155880672 | 0.19 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr2_+_162016804 | 0.18 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr13_+_34392185 | 0.18 |
ENST00000380071.3 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr12_-_57630873 | 0.17 |
ENST00000556732.1 |
NDUFA4L2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr12_+_15475331 | 0.16 |
ENST00000281171.4 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr10_+_52750930 | 0.16 |
ENST00000401604.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr14_+_22409308 | 0.15 |
ENST00000390441.2 |
TRAV9-2 |
T cell receptor alpha variable 9-2 |
| chr5_+_169010638 | 0.15 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr15_-_44069741 | 0.15 |
ENST00000319359.3 |
ELL3 |
elongation factor RNA polymerase II-like 3 |
| chr10_+_94352956 | 0.15 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr6_+_29141311 | 0.14 |
ENST00000377167.2 |
OR2J2 |
olfactory receptor, family 2, subfamily J, member 2 |
| chr3_-_126373929 | 0.14 |
ENST00000523403.1 ENST00000524230.2 |
TXNRD3 |
thioredoxin reductase 3 |
| chr5_-_169725231 | 0.14 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr17_-_56769382 | 0.14 |
ENST00000240361.8 ENST00000349033.5 ENST00000389934.3 |
TEX14 |
testis expressed 14 |
| chr3_+_100428268 | 0.14 |
ENST00000240851.4 |
TFG |
TRK-fused gene |
| chr3_-_125775629 | 0.13 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr3_+_100428316 | 0.13 |
ENST00000479672.1 ENST00000476228.1 ENST00000463568.1 |
TFG |
TRK-fused gene |
| chr14_+_96722539 | 0.13 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr11_-_85376121 | 0.13 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chr11_+_62037622 | 0.12 |
ENST00000227918.2 ENST00000525380.1 |
SCGB2A2 |
secretoglobin, family 2A, member 2 |
| chr5_+_140248518 | 0.12 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr3_+_100428188 | 0.12 |
ENST00000418917.2 ENST00000490574.1 |
TFG |
TRK-fused gene |
| chr10_-_69455873 | 0.11 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr1_+_55107449 | 0.11 |
ENST00000421030.2 ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7 |
maestro heat-like repeat family member 7 |
| chr19_-_6279932 | 0.11 |
ENST00000252674.7 |
MLLT1 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr2_+_162016916 | 0.11 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr22_+_32439019 | 0.11 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr15_+_85523671 | 0.11 |
ENST00000310298.4 ENST00000557957.1 |
PDE8A |
phosphodiesterase 8A |
| chr1_-_47407111 | 0.11 |
ENST00000371904.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr17_+_29664830 | 0.11 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr10_+_51549498 | 0.10 |
ENST00000358559.2 ENST00000298239.6 |
MSMB |
microseminoprotein, beta- |
| chr11_+_60995849 | 0.10 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr1_-_150208412 | 0.10 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_101569696 | 0.10 |
ENST00000597120.1 |
AC008948.1 |
AC008948.1 |
| chr1_+_17559776 | 0.09 |
ENST00000537499.1 ENST00000413717.2 ENST00000536552.1 |
PADI1 |
peptidyl arginine deiminase, type I |
| chr6_-_75953484 | 0.09 |
ENST00000472311.2 ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2 |
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr21_+_39628655 | 0.09 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr6_-_49681235 | 0.08 |
ENST00000339139.4 |
CRISP2 |
cysteine-rich secretory protein 2 |
| chr12_-_47473425 | 0.08 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr1_-_86174065 | 0.07 |
ENST00000370574.3 ENST00000431532.2 |
ZNHIT6 |
zinc finger, HIT-type containing 6 |
| chr6_-_146057144 | 0.07 |
ENST00000367519.3 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr12_-_114841703 | 0.07 |
ENST00000526441.1 |
TBX5 |
T-box 5 |
| chr2_+_89986318 | 0.07 |
ENST00000491977.1 |
IGKV2D-29 |
immunoglobulin kappa variable 2D-29 |
| chr1_-_55266926 | 0.06 |
ENST00000371276.4 |
TTC22 |
tetratricopeptide repeat domain 22 |
| chr19_+_21106081 | 0.06 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chr12_+_4758264 | 0.05 |
ENST00000266544.5 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr19_-_23869999 | 0.05 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr16_-_68033356 | 0.05 |
ENST00000393847.1 ENST00000573808.1 ENST00000572624.1 |
DPEP2 |
dipeptidase 2 |
| chr11_+_49050504 | 0.05 |
ENST00000332682.7 |
TRIM49B |
tripartite motif containing 49B |
| chr4_+_71248795 | 0.04 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr1_-_150208498 | 0.04 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_+_72052983 | 0.03 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr9_+_137979506 | 0.03 |
ENST00000539529.1 ENST00000392991.4 ENST00000371793.3 |
OLFM1 |
olfactomedin 1 |
| chr17_+_3118915 | 0.03 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chrX_-_100129128 | 0.03 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
| chr4_-_170533723 | 0.03 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr2_-_37384175 | 0.02 |
ENST00000411537.2 ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2 |
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr1_+_47603109 | 0.02 |
ENST00000371890.3 ENST00000294337.3 ENST00000371891.3 |
CYP4A22 |
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr2_+_172778952 | 0.02 |
ENST00000392584.1 ENST00000264108.4 |
HAT1 |
histone acetyltransferase 1 |
| chr4_+_110749143 | 0.02 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr4_+_106631966 | 0.02 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr17_-_39637392 | 0.02 |
ENST00000246639.2 ENST00000393989.1 |
KRT35 |
keratin 35 |
| chr5_-_64920115 | 0.02 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr6_-_33239612 | 0.02 |
ENST00000482399.1 ENST00000445902.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr17_-_39526052 | 0.02 |
ENST00000251646.3 |
KRT33B |
keratin 33B |
| chr17_-_3496171 | 0.02 |
ENST00000399756.4 |
TRPV1 |
transient receptor potential cation channel, subfamily V, member 1 |
| chr11_+_3011093 | 0.02 |
ENST00000332881.2 |
AC131971.1 |
HCG1782999; PRO0943; Uncharacterized protein |
| chr6_-_33239712 | 0.01 |
ENST00000436044.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr11_-_124670550 | 0.01 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr12_-_53207842 | 0.01 |
ENST00000458244.2 |
KRT4 |
keratin 4 |
| chr4_+_122722466 | 0.01 |
ENST00000243498.5 ENST00000379663.3 ENST00000509800.1 |
EXOSC9 |
exosome component 9 |
| chr20_+_36946029 | 0.01 |
ENST00000417318.1 |
BPI |
bactericidal/permeability-increasing protein |
| chr2_+_48796120 | 0.01 |
ENST00000394754.1 |
STON1-GTF2A1L |
STON1-GTF2A1L readthrough |
| chr17_-_46806540 | 0.00 |
ENST00000290295.7 |
HOXB13 |
homeobox B13 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 1.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 1.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 0.8 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 0.8 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.2 | 2.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 1.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 1.5 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 1.0 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.9 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 4.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.4 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 4.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.3 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.2 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 3.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 3.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.3 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.4 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.8 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 1.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 2.4 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.0 | 0.7 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 4.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 1.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 1.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 4.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 2.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 4.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 3.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.9 | GO:0003823 | antigen binding(GO:0003823) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 0.8 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.2 | 4.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 2.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 2.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 4.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 1.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 4.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |