Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HOXC9
Z-value: 0.66
Transcription factors associated with HOXC9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC9
|
ENSG00000180806.4 | HOXC9 |
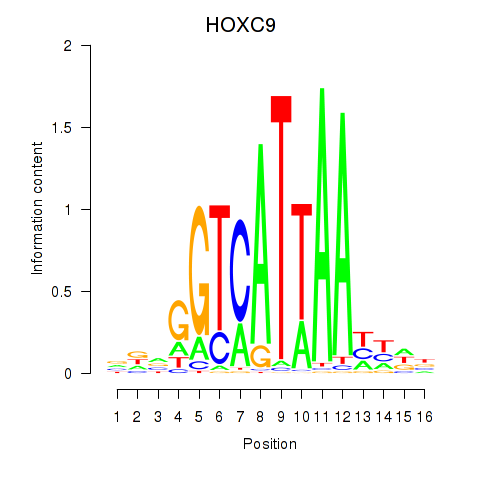
Activity profile of HOXC9 motif
Sorted Z-values of HOXC9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_173176452 | 2.24 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr6_-_52859046 | 1.96 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr7_+_130126165 | 1.24 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr12_-_91546926 | 1.18 |
ENST00000550758.1 |
DCN |
decorin |
| chr7_+_130126012 | 1.14 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr8_-_17555164 | 0.94 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr5_-_111093167 | 0.91 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr4_-_70626314 | 0.90 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr12_+_75874580 | 0.81 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr1_+_220863187 | 0.72 |
ENST00000294889.5 |
C1orf115 |
chromosome 1 open reading frame 115 |
| chr8_-_17579726 | 0.72 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr4_-_70626430 | 0.71 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chrX_+_15525426 | 0.71 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr18_+_32558208 | 0.69 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr3_+_138066539 | 0.64 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr1_+_163039143 | 0.63 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr11_+_19798964 | 0.63 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr5_-_16738451 | 0.60 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr6_+_83903061 | 0.57 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr16_-_12897642 | 0.56 |
ENST00000433677.2 ENST00000261660.4 ENST00000381774.4 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
| chr13_-_24007815 | 0.51 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr12_-_25055177 | 0.50 |
ENST00000538118.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr5_-_16916624 | 0.49 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr19_-_48752812 | 0.46 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr8_-_93029865 | 0.46 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_48753104 | 0.45 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr6_-_52859968 | 0.45 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr16_-_66764119 | 0.45 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr1_+_221051699 | 0.43 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr13_-_33760216 | 0.42 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr2_+_109204909 | 0.39 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr1_+_81771806 | 0.39 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr3_+_122785895 | 0.37 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr21_-_27423339 | 0.36 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr10_-_13344341 | 0.34 |
ENST00000396920.3 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
| chr11_+_114310102 | 0.32 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr12_-_30887948 | 0.32 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr2_+_37571845 | 0.31 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr2_+_109237717 | 0.31 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr2_+_37571717 | 0.31 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr5_-_10761206 | 0.30 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr7_+_80275621 | 0.30 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr5_-_42811986 | 0.29 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr2_+_162016916 | 0.29 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr11_+_114310164 | 0.29 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr11_+_5710919 | 0.27 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr7_+_80275752 | 0.27 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr16_+_22501658 | 0.26 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr5_-_42812143 | 0.26 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr11_+_71900572 | 0.26 |
ENST00000312293.4 |
FOLR1 |
folate receptor 1 (adult) |
| chr20_+_57414743 | 0.25 |
ENST00000313949.7 |
GNAS |
GNAS complex locus |
| chr20_-_33732952 | 0.25 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_+_131549610 | 0.25 |
ENST00000223865.8 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr2_+_109204743 | 0.25 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_-_111804905 | 0.24 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_+_153775821 | 0.24 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chrX_-_9734004 | 0.24 |
ENST00000467482.1 ENST00000380929.2 |
GPR143 |
G protein-coupled receptor 143 |
| chr2_+_162016804 | 0.24 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_-_86850949 | 0.24 |
ENST00000237455.4 |
RNF103 |
ring finger protein 103 |
| chr1_+_158979792 | 0.23 |
ENST00000359709.3 ENST00000430894.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr3_+_149191723 | 0.22 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr11_+_71900703 | 0.22 |
ENST00000393681.2 |
FOLR1 |
folate receptor 1 (adult) |
| chr12_-_90049878 | 0.22 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_202047596 | 0.22 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr12_-_90049828 | 0.22 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_+_71600144 | 0.22 |
ENST00000502653.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr2_+_109271481 | 0.22 |
ENST00000542845.1 ENST00000393314.2 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr7_-_22539771 | 0.21 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr2_-_70475701 | 0.21 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr10_+_45495898 | 0.21 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr7_+_80275953 | 0.21 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr2_+_161993412 | 0.20 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_115614370 | 0.20 |
ENST00000369301.3 |
NHLRC2 |
NHL repeat containing 2 |
| chr2_+_162016827 | 0.20 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr22_-_32651326 | 0.19 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr11_-_13517565 | 0.19 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr22_+_21133469 | 0.19 |
ENST00000406799.1 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr2_-_70475730 | 0.18 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr13_-_30424821 | 0.18 |
ENST00000380680.4 |
UBL3 |
ubiquitin-like 3 |
| chr4_+_69681710 | 0.18 |
ENST00000265403.7 ENST00000458688.2 |
UGT2B10 |
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr17_+_42264556 | 0.18 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr15_-_90222610 | 0.18 |
ENST00000300055.5 |
PLIN1 |
perilipin 1 |
| chr6_+_158733692 | 0.17 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr2_-_231989808 | 0.17 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr9_+_137979506 | 0.17 |
ENST00000539529.1 ENST00000392991.4 ENST00000371793.3 |
OLFM1 |
olfactomedin 1 |
| chr15_-_90222642 | 0.17 |
ENST00000430628.2 |
PLIN1 |
perilipin 1 |
| chr6_-_83903600 | 0.16 |
ENST00000506587.1 ENST00000507554.1 |
PGM3 |
phosphoglucomutase 3 |
| chr1_+_157963063 | 0.16 |
ENST00000360089.4 ENST00000368173.3 ENST00000392272.2 |
KIRREL |
kin of IRRE like (Drosophila) |
| chr1_+_35734562 | 0.16 |
ENST00000314607.6 ENST00000373297.2 |
ZMYM4 |
zinc finger, MYM-type 4 |
| chr2_+_202047843 | 0.15 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr5_-_125930929 | 0.15 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr19_-_48894104 | 0.15 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr16_-_66584059 | 0.14 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr9_+_131549483 | 0.13 |
ENST00000372648.5 ENST00000539497.1 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr7_-_44122063 | 0.13 |
ENST00000335195.6 ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM |
polymerase (DNA directed), mu |
| chr1_-_148347506 | 0.13 |
ENST00000369189.3 |
NBPF20 |
neuroblastoma breakpoint family, member 20 |
| chr17_+_68071389 | 0.13 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_147599549 | 0.13 |
ENST00000369228.5 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chrX_-_135338503 | 0.13 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chr2_+_161993465 | 0.12 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_-_98079965 | 0.12 |
ENST00000289081.3 |
FANCC |
Fanconi anemia, complementation group C |
| chr3_-_48130707 | 0.12 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr1_+_151735431 | 0.12 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr22_+_32149927 | 0.12 |
ENST00000437411.1 ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5 |
DEP domain containing 5 |
| chrX_-_130423200 | 0.11 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr19_-_46105411 | 0.11 |
ENST00000323040.4 ENST00000544371.1 |
GPR4 OPA3 |
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr19_+_21106081 | 0.11 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr15_+_45003675 | 0.11 |
ENST00000558401.1 ENST00000559916.1 ENST00000544417.1 |
B2M |
beta-2-microglobulin |
| chr17_-_40288449 | 0.11 |
ENST00000552162.1 ENST00000550504.1 |
RAB5C |
RAB5C, member RAS oncogene family |
| chr2_-_225811747 | 0.10 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr20_+_57414795 | 0.10 |
ENST00000371098.2 ENST00000371075.3 |
GNAS |
GNAS complex locus |
| chr16_+_71560023 | 0.10 |
ENST00000572450.1 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr16_+_71560154 | 0.10 |
ENST00000539698.3 |
CHST4 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr2_-_209010874 | 0.09 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr2_-_69664586 | 0.09 |
ENST00000303698.3 ENST00000394305.1 ENST00000410022.2 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr14_+_21359558 | 0.09 |
ENST00000304639.3 |
RNASE3 |
ribonuclease, RNase A family, 3 |
| chr4_+_70916119 | 0.09 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr17_+_68071458 | 0.09 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_+_24241969 | 0.09 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr10_+_74927875 | 0.09 |
ENST00000242505.6 |
FAM149B1 |
family with sequence similarity 149, member B1 |
| chr2_-_224467093 | 0.09 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr22_-_32767017 | 0.08 |
ENST00000400234.1 |
RFPL3S |
RFPL3 antisense |
| chr20_-_23860373 | 0.08 |
ENST00000304710.4 |
CST5 |
cystatin D |
| chr1_-_19426149 | 0.08 |
ENST00000429347.2 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
| chr4_+_155484155 | 0.08 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr4_+_123844225 | 0.08 |
ENST00000274008.4 |
SPATA5 |
spermatogenesis associated 5 |
| chr16_-_66583701 | 0.08 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr16_+_25228242 | 0.07 |
ENST00000219660.5 |
AQP8 |
aquaporin 8 |
| chr4_-_103749205 | 0.07 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr16_+_72090053 | 0.07 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr8_+_24241789 | 0.07 |
ENST00000256412.4 ENST00000538205.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr1_+_22962948 | 0.07 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr12_+_20963647 | 0.07 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr19_+_16940198 | 0.06 |
ENST00000248054.5 ENST00000596802.1 ENST00000379803.1 |
SIN3B |
SIN3 transcription regulator family member B |
| chr22_-_32766972 | 0.06 |
ENST00000382084.4 ENST00000382086.2 |
RFPL3S |
RFPL3 antisense |
| chr1_+_26798955 | 0.06 |
ENST00000361427.5 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
| chr5_+_175288631 | 0.06 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chrX_+_65382433 | 0.05 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr17_-_66951474 | 0.05 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chrX_-_68385274 | 0.05 |
ENST00000374584.3 ENST00000590146.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr19_+_55014085 | 0.05 |
ENST00000351841.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_+_20963632 | 0.05 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_103749105 | 0.05 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_68385354 | 0.05 |
ENST00000361478.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr9_-_21187598 | 0.05 |
ENST00000421715.1 |
IFNA4 |
interferon, alpha 4 |
| chr14_+_53173890 | 0.05 |
ENST00000445930.2 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr20_-_48782639 | 0.05 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr19_-_54850417 | 0.04 |
ENST00000291759.4 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr8_+_132916318 | 0.04 |
ENST00000254624.5 ENST00000522709.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr14_+_53173910 | 0.04 |
ENST00000606149.1 ENST00000555339.1 ENST00000556813.1 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr16_+_56485402 | 0.04 |
ENST00000566157.1 ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1 |
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr1_-_197115818 | 0.04 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr19_+_55014013 | 0.04 |
ENST00000301202.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_-_10959892 | 0.04 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr4_-_103749179 | 0.04 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749313 | 0.04 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_71248795 | 0.04 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr8_-_30670384 | 0.04 |
ENST00000221138.4 ENST00000518243.1 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr9_+_131062367 | 0.04 |
ENST00000601297.1 |
AL359091.2 |
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr1_-_226926864 | 0.04 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr8_-_110986918 | 0.04 |
ENST00000297404.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr2_+_27719697 | 0.04 |
ENST00000264717.2 ENST00000424318.2 |
GCKR |
glucokinase (hexokinase 4) regulator |
| chr7_-_36764004 | 0.04 |
ENST00000431169.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr6_-_146057144 | 0.03 |
ENST00000367519.3 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr17_+_33914276 | 0.03 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_148033693 | 0.03 |
ENST00000377888.3 ENST00000360693.3 |
HTR4 |
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr17_+_67498538 | 0.03 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr3_+_69812701 | 0.03 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr5_+_156696362 | 0.03 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_16370271 | 0.03 |
ENST00000375679.4 |
CLCNKB |
chloride channel, voltage-sensitive Kb |
| chr3_+_125687987 | 0.03 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr1_+_16348366 | 0.03 |
ENST00000375692.1 ENST00000420078.1 |
CLCNKA |
chloride channel, voltage-sensitive Ka |
| chr3_+_112709804 | 0.03 |
ENST00000383677.3 |
GTPBP8 |
GTP-binding protein 8 (putative) |
| chr17_+_45286706 | 0.03 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr16_+_28505955 | 0.02 |
ENST00000564831.1 ENST00000328423.5 ENST00000431282.1 |
APOBR |
apolipoprotein B receptor |
| chr1_+_16348497 | 0.02 |
ENST00000439316.2 |
CLCNKA |
chloride channel, voltage-sensitive Ka |
| chr3_-_38992052 | 0.02 |
ENST00000302328.3 ENST00000450244.1 ENST00000444237.2 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
| chr12_-_10605929 | 0.02 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_-_103748880 | 0.02 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr8_-_62602327 | 0.02 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr13_+_21714711 | 0.02 |
ENST00000607003.1 ENST00000492245.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr2_+_70121075 | 0.02 |
ENST00000409116.1 |
SNRNP27 |
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr15_-_41836441 | 0.02 |
ENST00000567866.1 ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1 |
RNA polymerase II associated protein 1 |
| chrX_-_73512177 | 0.02 |
ENST00000603672.1 ENST00000418855.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr4_+_70894130 | 0.02 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chrX_+_140982452 | 0.02 |
ENST00000544766.1 |
MAGEC3 |
melanoma antigen family C, 3 |
| chr4_+_155484103 | 0.02 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr2_+_233390890 | 0.02 |
ENST00000258385.3 ENST00000536614.1 ENST00000457943.2 |
CHRND |
cholinergic receptor, nicotinic, delta (muscle) |
| chr11_-_128894053 | 0.02 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr12_-_124118296 | 0.02 |
ENST00000424014.2 ENST00000537073.1 |
EIF2B1 |
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr14_+_72399833 | 0.02 |
ENST00000553530.1 ENST00000556437.1 |
RGS6 |
regulator of G-protein signaling 6 |
| chr12_-_11091862 | 0.02 |
ENST00000537503.1 |
TAS2R14 |
taste receptor, type 2, member 14 |
| chr2_+_106468204 | 0.02 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr20_+_56964169 | 0.02 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_+_58176525 | 0.01 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr6_-_22297730 | 0.01 |
ENST00000306482.1 |
PRL |
prolactin |
| chr11_-_114466471 | 0.01 |
ENST00000424261.2 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr17_+_7348658 | 0.01 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 0.6 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 1.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.5 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 1.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 0.6 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 1.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.5 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.2 | 0.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.8 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.5 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.3 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.2 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.4 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 2.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.3 | GO:0033216 | ferric iron import(GO:0033216) |
| 0.1 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 1.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 2.0 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |