Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
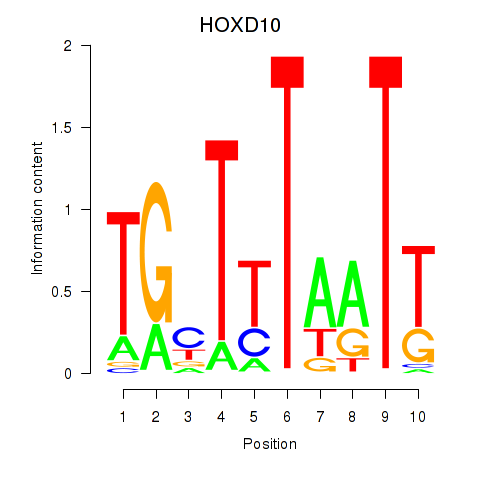
Results for HOXD10
Z-value: 0.73
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.5 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg19_v2_chr2_+_176981307_176981307 | -0.27 | 3.1e-01 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_32557610 | 2.91 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr6_-_32498046 | 2.11 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr13_-_47012325 | 1.92 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr11_+_60223225 | 1.90 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_-_111743285 | 1.90 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chr14_+_75988851 | 1.73 |
ENST00000555504.1 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr4_+_40198527 | 1.68 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr4_-_84035905 | 1.56 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr4_-_84035868 | 1.55 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr1_+_158978768 | 1.42 |
ENST00000447473.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_+_60223312 | 1.27 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_22689648 | 1.00 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr18_-_10701979 | 0.87 |
ENST00000538948.1 ENST00000285141.4 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr2_+_68592305 | 0.83 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr14_+_75988768 | 0.78 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr1_+_158801095 | 0.76 |
ENST00000368141.4 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr4_-_76944621 | 0.75 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr1_+_198608146 | 0.72 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr14_-_107049312 | 0.68 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr2_-_214016314 | 0.66 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr4_-_153332886 | 0.64 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chrX_+_37639264 | 0.63 |
ENST00000378588.4 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr18_+_29027696 | 0.62 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr1_+_79115503 | 0.60 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr1_+_241695670 | 0.60 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr8_-_79717750 | 0.59 |
ENST00000263851.4 ENST00000379113.2 |
IL7 |
interleukin 7 |
| chr11_-_104972158 | 0.59 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr11_-_104827425 | 0.56 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr4_-_76928641 | 0.56 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr1_+_174844645 | 0.54 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chrX_+_37639302 | 0.53 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chrX_-_154250989 | 0.50 |
ENST00000360256.4 |
F8 |
coagulation factor VIII, procoagulant component |
| chr11_-_104905840 | 0.43 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr7_+_90339169 | 0.42 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr1_+_186798073 | 0.40 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_+_61445007 | 0.40 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr10_+_35456444 | 0.39 |
ENST00000361599.4 |
CREM |
cAMP responsive element modulator |
| chr12_-_49333446 | 0.38 |
ENST00000537495.1 |
AC073610.5 |
Uncharacterized protein |
| chr16_-_24942273 | 0.37 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr2_-_89521942 | 0.36 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr8_+_124084899 | 0.35 |
ENST00000287380.1 ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31 |
TBC1 domain family, member 31 |
| chr19_-_51456321 | 0.35 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr15_+_25101698 | 0.35 |
ENST00000400097.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr1_-_197036364 | 0.34 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr1_-_156399184 | 0.33 |
ENST00000368243.1 ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr15_-_55562479 | 0.33 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chrX_-_148586804 | 0.33 |
ENST00000428056.2 ENST00000340855.6 ENST00000370441.4 ENST00000370443.4 |
IDS |
iduronate 2-sulfatase |
| chr12_+_16109519 | 0.33 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr4_-_90756769 | 0.33 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_-_26250835 | 0.32 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr14_-_106453155 | 0.31 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr10_+_120967072 | 0.29 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr5_-_150460914 | 0.29 |
ENST00000389378.2 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr2_+_166095898 | 0.29 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr14_+_37126765 | 0.29 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr11_+_122753391 | 0.28 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
| chr4_-_90758227 | 0.28 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_+_196313239 | 0.27 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr6_+_36097992 | 0.27 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chrX_+_15808569 | 0.26 |
ENST00000380308.3 ENST00000307771.7 |
ZRSR2 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr12_+_21525818 | 0.26 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr14_+_72064945 | 0.25 |
ENST00000537413.1 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr5_+_162930114 | 0.25 |
ENST00000280969.5 |
MAT2B |
methionine adenosyltransferase II, beta |
| chr17_-_67264947 | 0.24 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chrX_+_119737806 | 0.24 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chr13_+_114567131 | 0.23 |
ENST00000608651.1 |
GAS6-AS2 |
GAS6 antisense RNA 2 (head to head) |
| chr2_+_90198535 | 0.23 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr6_+_27791862 | 0.23 |
ENST00000355057.1 |
HIST1H4J |
histone cluster 1, H4j |
| chr10_+_11047259 | 0.22 |
ENST00000379261.4 ENST00000416382.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr9_+_42671887 | 0.22 |
ENST00000456520.1 ENST00000377391.3 |
CBWD7 |
COBW domain containing 7 |
| chr2_-_165424973 | 0.22 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr18_-_3845293 | 0.22 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_+_63953415 | 0.22 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr4_+_118955500 | 0.21 |
ENST00000296499.5 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr4_+_108910870 | 0.21 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr19_-_54618650 | 0.20 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr19_-_54619006 | 0.20 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr10_+_96522361 | 0.20 |
ENST00000371321.3 |
CYP2C19 |
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr9_-_36401155 | 0.20 |
ENST00000377885.2 |
RNF38 |
ring finger protein 38 |
| chr4_-_47983519 | 0.20 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr6_+_96025341 | 0.19 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr13_+_33160553 | 0.19 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr2_+_68961934 | 0.19 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr15_-_78538025 | 0.19 |
ENST00000560817.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_130569429 | 0.19 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr12_+_133195356 | 0.19 |
ENST00000389110.3 ENST00000449132.2 ENST00000343948.4 ENST00000352418.4 ENST00000350048.5 ENST00000351222.4 ENST00000348800.5 ENST00000542301.1 ENST00000536121.1 |
P2RX2 |
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr7_+_90338712 | 0.19 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr2_+_89998789 | 0.19 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr8_+_74903580 | 0.18 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr2_+_68961905 | 0.18 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr8_+_132952112 | 0.18 |
ENST00000520362.1 ENST00000519656.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr6_-_170101749 | 0.18 |
ENST00000448612.1 |
WDR27 |
WD repeat domain 27 |
| chr1_-_54405773 | 0.18 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr7_+_95115210 | 0.18 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr3_+_186435065 | 0.18 |
ENST00000287611.2 ENST00000265023.4 |
KNG1 |
kininogen 1 |
| chr10_+_13141441 | 0.18 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr9_+_27109133 | 0.18 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chrX_-_109590174 | 0.18 |
ENST00000372054.1 |
GNG5P2 |
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr1_-_92952433 | 0.17 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr18_-_3845321 | 0.17 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chrX_+_149887090 | 0.17 |
ENST00000538506.1 |
MTMR1 |
myotubularin related protein 1 |
| chr20_+_52105495 | 0.17 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr4_-_102268628 | 0.17 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_-_100016527 | 0.17 |
ENST00000523985.1 ENST00000518714.1 ENST00000520371.1 |
CCNC |
cyclin C |
| chr8_+_26150628 | 0.17 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_+_11679963 | 0.17 |
ENST00000263834.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr16_-_15736881 | 0.16 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr7_-_7782204 | 0.16 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chr17_+_29664830 | 0.16 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr22_-_38577731 | 0.16 |
ENST00000335539.3 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr5_+_140254884 | 0.15 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr16_+_30751953 | 0.15 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr2_-_90538397 | 0.15 |
ENST00000443397.3 |
RP11-685N3.1 |
Uncharacterized protein |
| chr6_+_5261225 | 0.15 |
ENST00000324331.6 |
FARS2 |
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr2_+_87565634 | 0.15 |
ENST00000421835.2 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr13_+_78109884 | 0.15 |
ENST00000377246.3 ENST00000349847.3 |
SCEL |
sciellin |
| chr18_-_61329118 | 0.15 |
ENST00000332821.8 ENST00000283752.5 |
SERPINB3 |
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr5_-_58882219 | 0.15 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr7_-_99698338 | 0.14 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr22_-_38577782 | 0.14 |
ENST00000430886.1 ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr10_+_96443378 | 0.14 |
ENST00000285979.6 |
CYP2C18 |
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr2_+_157330081 | 0.14 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr15_-_64673665 | 0.14 |
ENST00000300035.4 |
KIAA0101 |
KIAA0101 |
| chrX_-_110654147 | 0.13 |
ENST00000358070.4 |
DCX |
doublecortin |
| chr1_+_244214577 | 0.13 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr10_-_115614127 | 0.13 |
ENST00000369305.1 |
DCLRE1A |
DNA cross-link repair 1A |
| chr5_-_27038683 | 0.13 |
ENST00000511822.1 ENST00000231021.4 |
CDH9 |
cadherin 9, type 2 (T1-cadherin) |
| chr13_+_78109804 | 0.13 |
ENST00000535157.1 |
SCEL |
sciellin |
| chr1_+_74701062 | 0.13 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chrX_-_138724994 | 0.13 |
ENST00000536274.1 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr1_+_153963227 | 0.13 |
ENST00000368567.4 ENST00000392558.4 |
RPS27 |
ribosomal protein S27 |
| chr1_+_15986364 | 0.13 |
ENST00000345034.1 |
RSC1A1 |
regulatory solute carrier protein, family 1, member 1 |
| chr3_+_130569592 | 0.12 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_108541545 | 0.12 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_62439037 | 0.12 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr3_+_108541608 | 0.12 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_22233442 | 0.12 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_75784850 | 0.12 |
ENST00000550916.1 ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2 |
GLI pathogenesis-related 1 like 2 |
| chr5_+_118407053 | 0.12 |
ENST00000311085.8 ENST00000539542.1 |
DMXL1 |
Dmx-like 1 |
| chr18_-_3219847 | 0.12 |
ENST00000261606.7 |
MYOM1 |
myomesin 1 |
| chr12_-_56882136 | 0.11 |
ENST00000311966.4 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
| chr10_-_75226166 | 0.11 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr19_+_49588677 | 0.11 |
ENST00000598984.1 ENST00000598441.1 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
| chr1_-_13002348 | 0.11 |
ENST00000355096.2 |
PRAMEF6 |
PRAME family member 6 |
| chr2_-_161056762 | 0.11 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr1_-_40042073 | 0.11 |
ENST00000372858.3 |
PABPC4 |
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr6_-_24936170 | 0.11 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr13_-_50367057 | 0.11 |
ENST00000261667.3 |
KPNA3 |
karyopherin alpha 3 (importin alpha 4) |
| chr3_+_136649311 | 0.11 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr3_-_27498235 | 0.11 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_+_90273679 | 0.10 |
ENST00000423080.2 |
IGKV3D-7 |
immunoglobulin kappa variable 3D-7 |
| chr6_+_25754927 | 0.10 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr14_+_72399114 | 0.10 |
ENST00000553525.1 ENST00000555571.1 |
RGS6 |
regulator of G-protein signaling 6 |
| chr6_+_20534672 | 0.10 |
ENST00000274695.4 ENST00000378624.4 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
| chr8_-_117886955 | 0.10 |
ENST00000297338.2 |
RAD21 |
RAD21 homolog (S. pombe) |
| chr3_+_120461484 | 0.10 |
ENST00000484715.1 ENST00000469772.1 ENST00000283875.5 ENST00000492959.1 |
GTF2E1 |
general transcription factor IIE, polypeptide 1, alpha 56kDa |
| chr4_-_70725856 | 0.10 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr2_-_161056802 | 0.10 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr21_-_31588365 | 0.09 |
ENST00000399899.1 |
CLDN8 |
claudin 8 |
| chr2_-_190627481 | 0.09 |
ENST00000264151.5 ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1 |
O-sialoglycoprotein endopeptidase-like 1 |
| chr2_+_101437487 | 0.09 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr6_-_133055815 | 0.09 |
ENST00000509351.1 ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3 |
vanin 3 |
| chr7_-_115608304 | 0.09 |
ENST00000457268.1 |
TFEC |
transcription factor EC |
| chr6_-_5260963 | 0.09 |
ENST00000464010.1 ENST00000468929.1 ENST00000480566.1 |
LYRM4 |
LYR motif containing 4 |
| chr10_-_69455873 | 0.09 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr9_+_82187630 | 0.09 |
ENST00000265284.6 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_82187487 | 0.09 |
ENST00000435650.1 ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr22_-_50524298 | 0.09 |
ENST00000311597.5 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chrX_-_100872911 | 0.09 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr4_-_72649763 | 0.09 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr15_+_41549105 | 0.09 |
ENST00000560965.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr2_+_234590556 | 0.08 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr22_+_32871224 | 0.08 |
ENST00000452138.1 ENST00000382058.3 ENST00000397426.1 |
FBXO7 |
F-box protein 7 |
| chr19_+_15852203 | 0.08 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr1_+_14075903 | 0.08 |
ENST00000343137.4 ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr11_-_64647144 | 0.08 |
ENST00000359393.2 ENST00000433803.1 ENST00000411683.1 |
EHD1 |
EH-domain containing 1 |
| chr4_+_76649797 | 0.08 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr7_-_115670804 | 0.08 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr3_-_126327398 | 0.08 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr18_+_6729698 | 0.08 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr7_-_115670792 | 0.08 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr17_+_66624280 | 0.08 |
ENST00000585484.1 |
RP11-118B18.1 |
RP11-118B18.1 |
| chr3_+_186435137 | 0.08 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr3_+_138340049 | 0.08 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr18_+_21718924 | 0.08 |
ENST00000399496.3 |
CABYR |
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chrX_+_107334895 | 0.08 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr12_+_60083118 | 0.08 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_142967173 | 0.07 |
ENST00000370494.1 |
UBE2NL |
ubiquitin-conjugating enzyme E2N-like |
| chr3_-_195310802 | 0.07 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr5_+_131892603 | 0.07 |
ENST00000378823.3 ENST00000265335.6 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
| chr9_+_82188077 | 0.07 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_102861515 | 0.07 |
ENST00000262457.2 ENST00000541287.1 ENST00000262456.2 ENST00000374921.3 |
INVS |
inversin |
| chr18_+_3252265 | 0.07 |
ENST00000580887.1 ENST00000536605.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_149900122 | 0.07 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr4_+_187148556 | 0.07 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr11_-_102576537 | 0.07 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr8_-_101719159 | 0.07 |
ENST00000520868.1 ENST00000522658.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr6_-_5261141 | 0.07 |
ENST00000330636.4 ENST00000500576.2 |
LYRM4 |
LYR motif containing 4 |
| chr4_-_102268484 | 0.07 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_169629354 | 0.07 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.3 | 1.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 1.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 3.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.7 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.1 | 0.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 2.5 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.6 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.6 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 4.2 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 1.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 2.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0014848 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 1.5 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 5.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 3.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 6.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.8 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 2.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 2.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |