Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
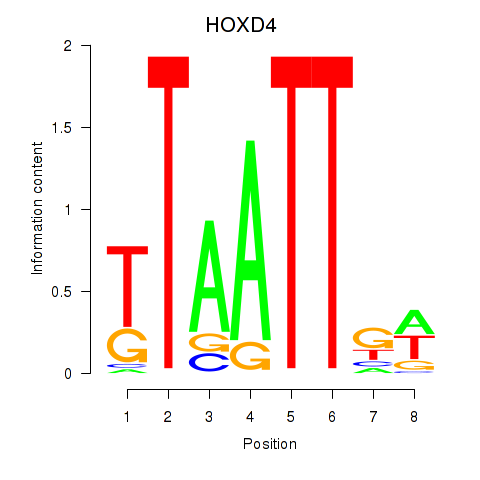
Results for HOXD4
Z-value: 0.80
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | HOXD4 |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_158787041 | 2.89 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr4_-_143226979 | 2.25 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr18_-_21891460 | 2.24 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr3_-_149095652 | 2.20 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr4_+_169753156 | 2.12 |
ENST00000393726.3 ENST00000507735.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr9_+_470288 | 2.03 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr4_+_169418195 | 1.98 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr4_-_143227088 | 1.66 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr11_+_101983176 | 1.59 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr13_+_73629107 | 1.37 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr18_+_29027696 | 1.31 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr4_+_169418255 | 1.17 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_+_186798073 | 1.16 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chrX_-_13835147 | 1.09 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr7_+_16793160 | 1.09 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chrX_-_10645773 | 1.06 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr8_-_18871159 | 1.01 |
ENST00000327040.8 ENST00000440756.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr1_+_144146808 | 0.92 |
ENST00000369190.5 ENST00000412624.2 ENST00000369365.3 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr2_-_165424973 | 0.90 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr11_+_12766583 | 0.88 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr6_+_53659746 | 0.87 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr2_-_31361543 | 0.86 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr3_-_149293990 | 0.80 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr3_-_126327398 | 0.61 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr17_-_10452929 | 0.59 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr12_-_91505608 | 0.55 |
ENST00000266718.4 |
LUM |
lumican |
| chr21_-_27423339 | 0.53 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr3_-_18480260 | 0.51 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr15_-_56209306 | 0.49 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr5_+_140743859 | 0.49 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr1_+_84630645 | 0.48 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_64225508 | 0.46 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_109237717 | 0.45 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr14_+_90863327 | 0.45 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr11_-_89224508 | 0.44 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr18_-_61311485 | 0.40 |
ENST00000436264.1 ENST00000356424.6 ENST00000341074.5 |
SERPINB4 |
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr2_-_190927447 | 0.39 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr5_+_140762268 | 0.39 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr11_-_89224638 | 0.37 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr4_+_155484155 | 0.37 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr2_+_234590556 | 0.36 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr4_+_70796784 | 0.36 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr11_-_89224139 | 0.35 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr4_+_155484103 | 0.34 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr11_-_89223883 | 0.34 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr8_-_27941380 | 0.33 |
ENST00000413272.2 ENST00000341513.6 |
NUGGC |
nuclear GTPase, germinal center associated |
| chr7_-_137028534 | 0.33 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr2_-_160473114 | 0.33 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr11_-_89224299 | 0.32 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224488 | 0.32 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr5_-_179499086 | 0.31 |
ENST00000261947.4 |
RNF130 |
ring finger protein 130 |
| chr12_+_26348246 | 0.31 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr5_-_179499108 | 0.30 |
ENST00000521389.1 |
RNF130 |
ring finger protein 130 |
| chrX_-_92928557 | 0.30 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr19_+_50016610 | 0.30 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_+_142468361 | 0.29 |
ENST00000367630.4 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
| chr17_+_66521936 | 0.29 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr12_+_16109519 | 0.28 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr1_-_235814048 | 0.27 |
ENST00000450593.1 ENST00000366598.4 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
| chr5_+_66300446 | 0.27 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr15_-_65809991 | 0.27 |
ENST00000559526.1 ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr3_-_124774802 | 0.23 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr2_+_204571375 | 0.23 |
ENST00000374478.4 |
CD28 |
CD28 molecule |
| chr17_-_73844722 | 0.23 |
ENST00000586257.1 |
WBP2 |
WW domain binding protein 2 |
| chr6_-_87804815 | 0.23 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr6_-_36515177 | 0.22 |
ENST00000229812.7 |
STK38 |
serine/threonine kinase 38 |
| chr7_-_137028498 | 0.20 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr5_-_137475071 | 0.20 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chr2_+_234826016 | 0.19 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_+_197518100 | 0.19 |
ENST00000438796.2 ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr5_+_101569696 | 0.18 |
ENST00000597120.1 |
AC008948.1 |
AC008948.1 |
| chr2_-_183291741 | 0.18 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr19_-_5903714 | 0.17 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr16_-_21663919 | 0.17 |
ENST00000569602.1 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr3_-_11623804 | 0.16 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr1_-_101360331 | 0.16 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr18_+_22040620 | 0.15 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr10_+_95848824 | 0.15 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr1_-_151762943 | 0.15 |
ENST00000368825.3 ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH |
tudor and KH domain containing |
| chr14_-_99947168 | 0.14 |
ENST00000331768.5 |
SETD3 |
SET domain containing 3 |
| chr2_-_207024233 | 0.13 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr11_-_64684672 | 0.12 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr2_-_99279928 | 0.12 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr1_-_87379785 | 0.12 |
ENST00000401030.3 ENST00000370554.1 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr2_+_207024306 | 0.11 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr18_+_22040593 | 0.11 |
ENST00000256906.4 |
HRH4 |
histamine receptor H4 |
| chr1_+_149754227 | 0.10 |
ENST00000444948.1 ENST00000369168.4 |
FCGR1A |
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chrX_+_37850026 | 0.09 |
ENST00000341016.3 |
CXorf27 |
chromosome X open reading frame 27 |
| chr17_-_29624343 | 0.09 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr2_+_171034646 | 0.09 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr18_-_51750948 | 0.09 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr5_-_55412774 | 0.08 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr2_-_166060571 | 0.08 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr18_-_19994830 | 0.07 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr3_+_142342228 | 0.07 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr2_+_88047606 | 0.06 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr14_-_101036119 | 0.06 |
ENST00000355173.2 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr6_-_136847099 | 0.06 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr1_-_36948879 | 0.06 |
ENST00000373106.1 ENST00000373104.1 ENST00000373103.1 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr3_-_195310802 | 0.05 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr15_-_88799661 | 0.05 |
ENST00000360948.2 ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr8_-_72274467 | 0.05 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr5_+_179921344 | 0.04 |
ENST00000261951.4 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr2_-_166060552 | 0.04 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_-_170744498 | 0.04 |
ENST00000382808.4 ENST00000314251.3 |
SLC2A2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr14_-_24911971 | 0.04 |
ENST00000555365.1 ENST00000399395.3 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr20_+_18447771 | 0.04 |
ENST00000377603.4 |
POLR3F |
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr14_+_23067146 | 0.04 |
ENST00000428304.2 |
ABHD4 |
abhydrolase domain containing 4 |
| chr17_+_3118915 | 0.03 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chr4_-_46996424 | 0.03 |
ENST00000264318.3 |
GABRA4 |
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr15_+_80364901 | 0.03 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr17_+_41132564 | 0.02 |
ENST00000361677.1 ENST00000589705.1 |
RUNDC1 |
RUN domain containing 1 |
| chr2_-_160472952 | 0.02 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr14_-_64761078 | 0.02 |
ENST00000341099.4 ENST00000556275.1 ENST00000542956.1 ENST00000353772.3 ENST00000357782.2 ENST00000267525.6 |
ESR2 |
estrogen receptor 2 (ER beta) |
| chr14_-_24911868 | 0.02 |
ENST00000554698.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr16_-_21663950 | 0.02 |
ENST00000268389.4 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr2_-_228497888 | 0.02 |
ENST00000264387.4 ENST00000409066.1 |
C2orf83 |
chromosome 2 open reading frame 83 |
| chr12_-_91451758 | 0.01 |
ENST00000266719.3 |
KERA |
keratocan |
| chr1_-_120935894 | 0.01 |
ENST00000369383.4 ENST00000369384.4 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr19_+_48828582 | 0.01 |
ENST00000270221.6 ENST00000596315.1 |
EMP3 |
epithelial membrane protein 3 |
| chr15_-_50558223 | 0.01 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr10_-_17171817 | 0.00 |
ENST00000377833.4 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
| chr11_-_77790865 | 0.00 |
ENST00000534029.1 ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2 NDUFC2-KCTD14 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr12_+_9980113 | 0.00 |
ENST00000537723.1 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
| chr8_+_24151620 | 0.00 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr11_-_104972158 | 0.00 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 5.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.5 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 1.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 1.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.5 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 1.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 2.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.2 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.1 | 0.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.1 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 3.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.5 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.9 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 5.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 2.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0034236 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 5.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 3.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |