Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
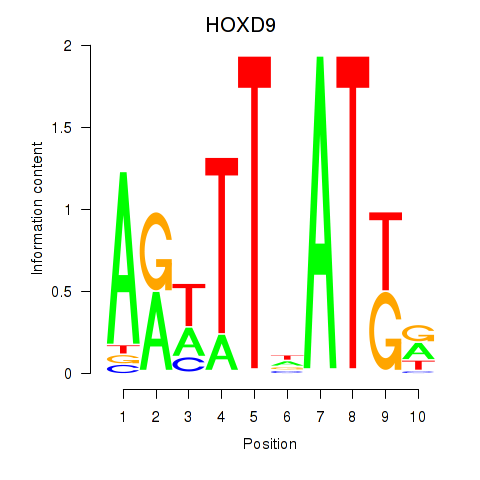
Results for HOXD9
Z-value: 1.08
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | HOXD9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.32 | 2.3e-01 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_49294472 | 2.97 |
ENST00000361446.5 |
GAGE12B |
G antigen 12B |
| chr6_+_6588316 | 2.82 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr2_+_143635067 | 2.72 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr2_+_143886877 | 2.33 |
ENST00000295095.6 |
ARHGAP15 |
Rho GTPase activating protein 15 |
| chr6_+_13272904 | 2.25 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr12_-_15104040 | 2.24 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_158345462 | 2.15 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr1_+_198608146 | 2.06 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr19_+_10397648 | 2.01 |
ENST00000340992.4 ENST00000393717.2 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chrX_-_52258669 | 1.80 |
ENST00000441417.1 |
XAGE1A |
X antigen family, member 1A |
| chr11_-_5255861 | 1.77 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr13_-_99959641 | 1.67 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr2_+_33661382 | 1.64 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr12_-_8765446 | 1.53 |
ENST00000537228.1 ENST00000229335.6 |
AICDA |
activation-induced cytidine deaminase |
| chrX_+_52513455 | 1.52 |
ENST00000446098.1 |
XAGE1C |
X antigen family, member 1C |
| chr13_-_47012325 | 1.44 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr11_-_104972158 | 1.41 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr15_-_55562582 | 1.31 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr6_-_32634425 | 1.27 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr2_-_170430277 | 1.27 |
ENST00000438035.1 ENST00000453929.2 |
FASTKD1 |
FAST kinase domains 1 |
| chr2_-_170430366 | 1.23 |
ENST00000453153.2 ENST00000445210.1 |
FASTKD1 |
FAST kinase domains 1 |
| chr10_-_115614127 | 1.22 |
ENST00000369305.1 |
DCLRE1A |
DNA cross-link repair 1A |
| chr6_+_32812568 | 1.14 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_-_76944621 | 1.04 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr14_+_39583427 | 1.04 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr17_-_29641084 | 1.03 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr8_+_86089460 | 0.99 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr8_+_86089619 | 0.95 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr2_+_143635222 | 0.86 |
ENST00000375773.2 ENST00000409512.1 ENST00000410015.2 |
KYNU |
kynureninase |
| chr1_-_39339777 | 0.80 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr4_-_144940477 | 0.79 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr2_+_191334212 | 0.78 |
ENST00000444317.1 ENST00000535751.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr2_+_90198535 | 0.77 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr4_-_144826682 | 0.73 |
ENST00000358615.4 ENST00000437468.2 |
GYPE |
glycophorin E (MNS blood group) |
| chr7_-_5998714 | 0.73 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr12_-_118628350 | 0.72 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr13_+_33160553 | 0.72 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr4_-_84035868 | 0.72 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr15_+_66797627 | 0.71 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr15_+_66797455 | 0.69 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr2_-_176046391 | 0.69 |
ENST00000392541.3 ENST00000409194.1 |
ATP5G3 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_-_241799232 | 0.68 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr3_+_178276488 | 0.65 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr17_-_47723943 | 0.64 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chr3_+_182511266 | 0.63 |
ENST00000323116.5 ENST00000493826.1 |
ATP11B |
ATPase, class VI, type 11B |
| chr6_+_24775153 | 0.62 |
ENST00000356509.3 ENST00000230056.3 |
GMNN |
geminin, DNA replication inhibitor |
| chr2_-_191115229 | 0.61 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chrX_-_33229636 | 0.58 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr13_+_51483814 | 0.56 |
ENST00000336617.3 ENST00000422660.1 |
RNASEH2B |
ribonuclease H2, subunit B |
| chr4_-_84035905 | 0.56 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chrX_-_110513703 | 0.54 |
ENST00000324068.1 |
CAPN6 |
calpain 6 |
| chr11_-_104905840 | 0.53 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chrX_+_149887090 | 0.52 |
ENST00000538506.1 |
MTMR1 |
myotubularin related protein 1 |
| chr17_+_7155819 | 0.49 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr11_-_128457446 | 0.49 |
ENST00000392668.4 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr8_-_86253888 | 0.48 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr14_-_106725723 | 0.47 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr18_-_74728998 | 0.45 |
ENST00000359645.3 ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP |
myelin basic protein |
| chr1_+_95616933 | 0.43 |
ENST00000604203.1 |
RP11-57H12.6 |
TMEM56-RWDD3 readthrough |
| chr7_+_77469439 | 0.43 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chrX_+_118602363 | 0.43 |
ENST00000317881.8 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr19_-_56826157 | 0.43 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr4_-_76928641 | 0.43 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr17_+_58018269 | 0.42 |
ENST00000591035.1 |
RP11-178C3.1 |
Uncharacterized protein |
| chr6_-_28220002 | 0.42 |
ENST00000377294.2 |
ZKSCAN4 |
zinc finger with KRAB and SCAN domains 4 |
| chrX_-_72434628 | 0.42 |
ENST00000536638.1 ENST00000373517.3 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
| chr17_-_46690839 | 0.40 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr11_+_92085262 | 0.40 |
ENST00000298047.6 ENST00000409404.2 ENST00000541502.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr21_-_19775973 | 0.40 |
ENST00000284885.3 |
TMPRSS15 |
transmembrane protease, serine 15 |
| chr19_+_52800410 | 0.39 |
ENST00000595962.1 ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480 |
zinc finger protein 480 |
| chr1_-_197036364 | 0.38 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr19_-_53466095 | 0.37 |
ENST00000391786.2 ENST00000434371.2 ENST00000357666.4 ENST00000438970.2 ENST00000270457.4 ENST00000535506.1 ENST00000444460.2 ENST00000457013.2 |
ZNF816 |
zinc finger protein 816 |
| chr6_-_133055815 | 0.36 |
ENST00000509351.1 ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3 |
vanin 3 |
| chr9_+_70856397 | 0.36 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr7_+_120629653 | 0.36 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr18_-_57027194 | 0.34 |
ENST00000251047.5 |
LMAN1 |
lectin, mannose-binding, 1 |
| chr5_-_78808617 | 0.33 |
ENST00000282260.6 ENST00000508576.1 ENST00000535690.1 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr6_+_118869452 | 0.32 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr9_-_21228221 | 0.32 |
ENST00000413767.2 |
IFNA17 |
interferon, alpha 17 |
| chr21_+_35736302 | 0.32 |
ENST00000290310.3 |
KCNE2 |
potassium voltage-gated channel, Isk-related family, member 2 |
| chr17_-_73389737 | 0.32 |
ENST00000392563.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr4_+_118955500 | 0.31 |
ENST00000296499.5 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_-_236046872 | 0.31 |
ENST00000536965.1 |
LYST |
lysosomal trafficking regulator |
| chr2_-_225434538 | 0.31 |
ENST00000409096.1 |
CUL3 |
cullin 3 |
| chrX_+_120181457 | 0.28 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr3_-_107777208 | 0.28 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr1_+_44870866 | 0.28 |
ENST00000355387.2 ENST00000361799.2 |
RNF220 |
ring finger protein 220 |
| chr2_+_114647504 | 0.28 |
ENST00000263238.2 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
| chr5_-_96518907 | 0.28 |
ENST00000508447.1 ENST00000283109.3 |
RIOK2 |
RIO kinase 2 |
| chr17_-_39191107 | 0.27 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chrX_+_12993336 | 0.26 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr15_+_52155001 | 0.26 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr8_+_104892639 | 0.26 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr14_+_65878565 | 0.26 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr5_-_41261540 | 0.26 |
ENST00000263413.3 |
C6 |
complement component 6 |
| chr2_-_228244013 | 0.25 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chr4_+_69313145 | 0.24 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr5_+_140207536 | 0.23 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr9_+_12693336 | 0.22 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr6_+_10528560 | 0.22 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr2_+_210444748 | 0.22 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chrX_+_135730373 | 0.21 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr6_+_63921351 | 0.20 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr17_-_39646116 | 0.20 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr5_-_138775177 | 0.20 |
ENST00000302060.5 |
DNAJC18 |
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr21_-_31588365 | 0.20 |
ENST00000399899.1 |
CLDN8 |
claudin 8 |
| chr22_-_19466683 | 0.20 |
ENST00000399523.1 ENST00000421968.2 ENST00000447868.1 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr8_-_80942061 | 0.20 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr12_-_21927736 | 0.19 |
ENST00000240662.2 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr4_+_88720698 | 0.19 |
ENST00000226284.5 |
IBSP |
integrin-binding sialoprotein |
| chr2_+_185463093 | 0.19 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr5_+_140248518 | 0.19 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr12_+_58176525 | 0.18 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr12_+_15125954 | 0.17 |
ENST00000266395.2 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr1_+_197237352 | 0.17 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr8_-_117886955 | 0.17 |
ENST00000297338.2 |
RAD21 |
RAD21 homolog (S. pombe) |
| chr12_-_118797475 | 0.17 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr12_+_9980069 | 0.16 |
ENST00000354855.3 ENST00000324214.4 ENST00000279544.3 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
| chr7_+_93535866 | 0.16 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_+_57998923 | 0.15 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr19_+_54619125 | 0.15 |
ENST00000445811.1 ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31 |
pre-mRNA processing factor 31 |
| chr18_+_21452804 | 0.15 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr7_-_38289173 | 0.15 |
ENST00000436911.2 |
TRGC2 |
T cell receptor gamma constant 2 |
| chr15_+_58430368 | 0.14 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr8_-_20040601 | 0.14 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr4_+_71263599 | 0.14 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr4_+_71296204 | 0.14 |
ENST00000413702.1 |
MUC7 |
mucin 7, secreted |
| chrX_+_135730297 | 0.14 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr16_+_4545853 | 0.14 |
ENST00000575129.1 ENST00000398595.3 ENST00000414777.1 |
HMOX2 |
heme oxygenase (decycling) 2 |
| chr7_-_80141328 | 0.14 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr10_-_105845674 | 0.13 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr22_-_19466732 | 0.13 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr3_-_116163830 | 0.13 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chrX_+_99839799 | 0.13 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr1_-_152332480 | 0.13 |
ENST00000388718.5 |
FLG2 |
filaggrin family member 2 |
| chr2_+_145780739 | 0.13 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr3_+_145782358 | 0.12 |
ENST00000422482.1 |
AC107021.1 |
HCG1786590; PRO2533; Uncharacterized protein |
| chr6_+_46761118 | 0.11 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr8_-_20040638 | 0.11 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr11_+_49050504 | 0.11 |
ENST00000332682.7 |
TRIM49B |
tripartite motif containing 49B |
| chr10_+_98064085 | 0.11 |
ENST00000419175.1 ENST00000371174.2 |
DNTT |
DNA nucleotidylexotransferase |
| chr11_+_69924639 | 0.11 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr11_+_63606373 | 0.11 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr11_+_12115543 | 0.10 |
ENST00000537344.1 ENST00000532179.1 ENST00000526065.1 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr10_-_13523073 | 0.10 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chrY_+_15815447 | 0.10 |
ENST00000284856.3 |
TMSB4Y |
thymosin beta 4, Y-linked |
| chr4_+_71108300 | 0.09 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr7_+_93535817 | 0.09 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr9_-_21239978 | 0.09 |
ENST00000380222.2 |
IFNA14 |
interferon, alpha 14 |
| chr5_-_39274617 | 0.09 |
ENST00000510188.1 |
FYB |
FYN binding protein |
| chr10_+_17794251 | 0.08 |
ENST00000377495.1 ENST00000338221.5 |
TMEM236 |
transmembrane protein 236 |
| chr6_+_72926145 | 0.08 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr9_-_215744 | 0.08 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr1_+_67632083 | 0.08 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chr1_+_16083154 | 0.08 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr15_+_58430567 | 0.08 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr9_+_124103625 | 0.07 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr3_-_195310802 | 0.07 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr16_-_20556492 | 0.07 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr4_-_168155577 | 0.07 |
ENST00000541354.1 ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_+_25015968 | 0.07 |
ENST00000380834.2 ENST00000473706.1 |
CENPO |
centromere protein O |
| chr12_+_9144626 | 0.06 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr12_-_11002063 | 0.06 |
ENST00000544994.1 ENST00000228811.4 ENST00000540107.1 |
PRR4 |
proline rich 4 (lacrimal) |
| chr1_-_165414414 | 0.06 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr1_-_67266939 | 0.05 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr11_+_61957687 | 0.05 |
ENST00000306238.3 |
SCGB1D1 |
secretoglobin, family 1D, member 1 |
| chr2_+_145780767 | 0.05 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr4_-_68749745 | 0.05 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr2_-_228497888 | 0.05 |
ENST00000264387.4 ENST00000409066.1 |
C2orf83 |
chromosome 2 open reading frame 83 |
| chr6_-_49712123 | 0.05 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr3_+_155838337 | 0.05 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_173686303 | 0.04 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_45390222 | 0.04 |
ENST00000359524.5 |
RUNX2 |
runt-related transcription factor 2 |
| chr5_-_147211226 | 0.04 |
ENST00000296695.5 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr4_-_68749699 | 0.04 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr3_-_167191814 | 0.04 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chrX_-_109590174 | 0.04 |
ENST00000372054.1 |
GNG5P2 |
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr14_+_81421355 | 0.04 |
ENST00000541158.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr11_+_30253410 | 0.04 |
ENST00000533718.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr18_-_24443151 | 0.03 |
ENST00000440832.3 |
AQP4 |
aquaporin 4 |
| chr2_+_234668894 | 0.03 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_-_59043166 | 0.02 |
ENST00000371225.2 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
| chr18_-_5396271 | 0.02 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr4_+_88529681 | 0.02 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr2_+_1418154 | 0.01 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr12_-_11463353 | 0.01 |
ENST00000279575.1 ENST00000535904.1 ENST00000445719.2 |
PRB4 |
proline-rich protein BstNI subfamily 4 |
| chr18_-_53069419 | 0.01 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr1_+_181057638 | 0.01 |
ENST00000367577.4 |
IER5 |
immediate early response 5 |
| chr4_-_168155700 | 0.01 |
ENST00000357545.4 ENST00000512648.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_73076432 | 0.01 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr11_-_102401469 | 0.01 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr14_+_22615942 | 0.01 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr5_+_135496675 | 0.01 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr15_-_66797172 | 0.00 |
ENST00000569438.1 ENST00000569696.1 ENST00000307961.6 |
RPL4 |
ribosomal protein L4 |
| chr6_-_74231444 | 0.00 |
ENST00000331523.2 ENST00000356303.2 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 2.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 1.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.4 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 1.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 2.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 2.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.0 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.6 | 2.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.6 | 1.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.5 | 1.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 2.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.3 | 2.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.5 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 1.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.3 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0015722 | purine nucleobase transport(GO:0006863) canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 1.6 | GO:0002381 | immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0050817 | blood coagulation(GO:0007596) hemostasis(GO:0007599) coagulation(GO:0050817) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 2.3 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 2.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 4.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 0.3 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 3.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |