Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for HSF1
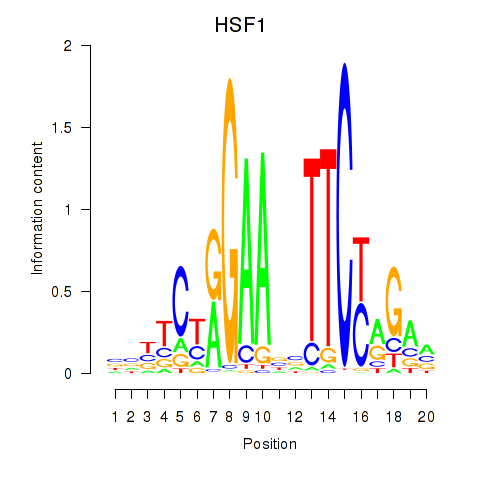
Z-value: 1.90
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | HSF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | 0.46 | 7.4e-02 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_111415757 | 5.95 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr6_-_33041378 | 4.16 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chrX_+_52511925 | 3.63 |
ENST00000375588.1 |
XAGE1C |
X antigen family, member 1C |
| chrX_-_52260199 | 3.40 |
ENST00000375600.1 |
XAGE1A |
X antigen family, member 1A |
| chr15_-_55581954 | 3.00 |
ENST00000336787.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_-_52911718 | 2.83 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr8_-_81083890 | 2.82 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chrX_-_154563889 | 2.76 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr14_-_106573756 | 2.58 |
ENST00000390601.2 |
IGHV3-11 |
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr18_+_57567180 | 2.56 |
ENST00000316660.6 ENST00000269518.9 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
| chrX_-_52533139 | 2.49 |
ENST00000374959.3 |
XAGE1D |
X antigen family, member 1D |
| chr14_-_106926724 | 2.45 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chrX_-_52546033 | 2.40 |
ENST00000375567.3 |
XAGE1E |
X antigen family, member 1E |
| chr19_-_7766991 | 2.39 |
ENST00000597921.1 ENST00000346664.5 |
FCER2 |
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chrX_-_73072534 | 2.34 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr19_-_51504411 | 2.33 |
ENST00000593490.1 |
KLK8 |
kallikrein-related peptidase 8 |
| chr2_+_102624977 | 2.31 |
ENST00000441002.1 |
IL1R2 |
interleukin 1 receptor, type II |
| chr15_-_55563072 | 2.24 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr8_-_81083341 | 2.24 |
ENST00000519303.2 |
TPD52 |
tumor protein D52 |
| chr20_+_57430162 | 2.24 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr12_+_93772402 | 2.08 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr6_+_32605195 | 2.08 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr5_+_118690466 | 2.06 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_32605134 | 2.04 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_-_32498046 | 2.04 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr19_-_42806444 | 2.00 |
ENST00000594989.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr6_-_32557610 | 1.92 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chrX_+_52238974 | 1.89 |
ENST00000375613.3 |
XAGE1B |
X antigen family, member 1B |
| chrX_+_30233668 | 1.89 |
ENST00000378988.4 |
MAGEB2 |
melanoma antigen family B, 2 |
| chrX_+_48380205 | 1.86 |
ENST00000446158.1 ENST00000414061.1 |
EBP |
emopamil binding protein (sterol isomerase) |
| chr18_+_21452964 | 1.83 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr14_-_106092403 | 1.81 |
ENST00000390543.2 |
IGHG4 |
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr6_+_89674246 | 1.80 |
ENST00000369474.1 |
AL079342.1 |
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr12_+_2904102 | 1.80 |
ENST00000001008.4 |
FKBP4 |
FK506 binding protein 4, 59kDa |
| chr17_-_39928106 | 1.77 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr18_+_21452804 | 1.71 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr1_-_153363452 | 1.71 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr19_-_42806723 | 1.70 |
ENST00000262890.3 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr16_+_28943260 | 1.68 |
ENST00000538922.1 ENST00000324662.3 ENST00000567541.1 |
CD19 |
CD19 molecule |
| chr19_-_42806919 | 1.62 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr16_+_85942594 | 1.62 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr1_+_35247859 | 1.59 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr8_+_142402089 | 1.57 |
ENST00000521578.1 ENST00000520105.1 ENST00000523147.1 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
| chr15_-_58357932 | 1.57 |
ENST00000347587.3 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr14_-_106668095 | 1.56 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr19_-_42806842 | 1.56 |
ENST00000596265.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr6_+_4890226 | 1.55 |
ENST00000343762.5 |
CDYL |
chromodomain protein, Y-like |
| chr17_-_39677971 | 1.53 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr6_-_41909466 | 1.52 |
ENST00000414200.2 |
CCND3 |
cyclin D3 |
| chr19_-_51522955 | 1.50 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr7_-_150652924 | 1.50 |
ENST00000330883.4 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr16_+_222846 | 1.50 |
ENST00000251595.6 ENST00000397806.1 |
HBA2 |
hemoglobin, alpha 2 |
| chr14_-_106552755 | 1.47 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr21_+_35445827 | 1.46 |
ENST00000608209.1 ENST00000381151.3 |
SLC5A3 SLC5A3 |
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr6_-_41909561 | 1.43 |
ENST00000372991.4 |
CCND3 |
cyclin D3 |
| chr5_-_141016382 | 1.39 |
ENST00000523088.1 ENST00000305264.3 |
HDAC3 |
histone deacetylase 3 |
| chr2_+_47596287 | 1.39 |
ENST00000263735.4 |
EPCAM |
epithelial cell adhesion molecule |
| chrX_-_48776292 | 1.36 |
ENST00000376509.4 |
PIM2 |
pim-2 oncogene |
| chr6_-_32784687 | 1.34 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr1_-_17215868 | 1.33 |
ENST00000422124.1 |
RP11-108M9.4 |
RP11-108M9.4 |
| chr2_-_191878162 | 1.29 |
ENST00000540176.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr19_+_782755 | 1.29 |
ENST00000606242.1 ENST00000586061.1 |
AC006273.5 |
AC006273.5 |
| chr18_-_19284724 | 1.26 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr19_-_51456198 | 1.26 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chrX_-_72434628 | 1.25 |
ENST00000536638.1 ENST00000373517.3 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
| chr15_+_78857870 | 1.25 |
ENST00000559554.1 |
CHRNA5 |
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr11_+_10471836 | 1.24 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr6_+_26597155 | 1.23 |
ENST00000274849.1 |
ABT1 |
activator of basal transcription 1 |
| chr20_-_57617831 | 1.23 |
ENST00000371033.5 ENST00000355937.4 |
SLMO2 |
slowmo homolog 2 (Drosophila) |
| chr5_+_122110691 | 1.22 |
ENST00000379516.2 ENST00000505934.1 ENST00000514949.1 |
SNX2 |
sorting nexin 2 |
| chr8_-_81083731 | 1.21 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr1_-_17216109 | 1.20 |
ENST00000416869.1 |
RP11-108M9.4 |
RP11-108M9.4 |
| chr2_-_9143786 | 1.20 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr19_-_2051223 | 1.19 |
ENST00000309340.7 ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr6_+_35265586 | 1.19 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr14_-_106518922 | 1.16 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr15_-_58357866 | 1.15 |
ENST00000537372.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr6_-_41909191 | 1.14 |
ENST00000512426.1 ENST00000372987.4 |
CCND3 |
cyclin D3 |
| chr19_-_51456344 | 1.13 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr15_+_81589254 | 1.13 |
ENST00000394652.2 |
IL16 |
interleukin 16 |
| chr4_-_25865159 | 1.12 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_+_20385231 | 1.12 |
ENST00000530266.1 ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr4_-_84030996 | 1.11 |
ENST00000411416.2 |
PLAC8 |
placenta-specific 8 |
| chrX_+_108779870 | 1.09 |
ENST00000372107.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr1_+_152957707 | 1.08 |
ENST00000368762.1 |
SPRR1A |
small proline-rich protein 1A |
| chr12_+_93772326 | 1.07 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr1_+_6105974 | 1.06 |
ENST00000378083.3 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr11_-_104905840 | 1.01 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr2_-_191878681 | 0.99 |
ENST00000409465.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr14_+_75988768 | 0.98 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr18_+_20513278 | 0.98 |
ENST00000327155.5 |
RBBP8 |
retinoblastoma binding protein 8 |
| chr19_+_1065922 | 0.97 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr8_-_57359131 | 0.97 |
ENST00000518974.1 ENST00000523051.1 ENST00000518770.1 ENST00000451791.2 |
PENK |
proenkephalin |
| chr16_+_68119764 | 0.97 |
ENST00000570212.1 ENST00000562926.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr11_+_60050026 | 0.95 |
ENST00000395016.3 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr11_-_47870091 | 0.95 |
ENST00000526870.1 |
NUP160 |
nucleoporin 160kDa |
| chr2_-_191885686 | 0.93 |
ENST00000432058.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr6_+_44215603 | 0.92 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr17_+_53828333 | 0.92 |
ENST00000268896.5 |
PCTP |
phosphatidylcholine transfer protein |
| chr1_+_28562617 | 0.90 |
ENST00000497986.1 ENST00000335514.5 ENST00000468425.2 ENST00000465645.1 |
ATPIF1 |
ATPase inhibitory factor 1 |
| chr2_+_86426478 | 0.90 |
ENST00000254644.8 ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35 |
mitochondrial ribosomal protein L35 |
| chr11_+_46354455 | 0.89 |
ENST00000343674.6 |
DGKZ |
diacylglycerol kinase, zeta |
| chr3_+_183853052 | 0.89 |
ENST00000273783.3 ENST00000432569.1 ENST00000444495.1 |
EIF2B5 |
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr5_+_32531893 | 0.88 |
ENST00000512913.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr15_-_60771128 | 0.88 |
ENST00000558512.1 ENST00000561114.1 |
NARG2 |
NMDA receptor regulated 2 |
| chr1_-_225615599 | 0.88 |
ENST00000421383.1 ENST00000272163.4 |
LBR |
lamin B receptor |
| chr6_-_167369612 | 0.88 |
ENST00000507747.1 |
RP11-514O12.4 |
RP11-514O12.4 |
| chr5_+_7654057 | 0.87 |
ENST00000537121.1 |
ADCY2 |
adenylate cyclase 2 (brain) |
| chrX_+_108780062 | 0.87 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chrX_-_100662881 | 0.87 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr7_-_150020578 | 0.84 |
ENST00000478393.1 |
ACTR3C |
ARP3 actin-related protein 3 homolog C (yeast) |
| chr16_+_19125252 | 0.82 |
ENST00000566735.1 ENST00000381440.3 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr2_-_101034070 | 0.82 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr15_+_78857849 | 0.81 |
ENST00000299565.5 |
CHRNA5 |
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr7_-_150754935 | 0.80 |
ENST00000297518.4 |
CDK5 |
cyclin-dependent kinase 5 |
| chr19_-_51523412 | 0.79 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr18_-_51751132 | 0.78 |
ENST00000256429.3 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr17_+_53828381 | 0.78 |
ENST00000576183.1 |
PCTP |
phosphatidylcholine transfer protein |
| chr11_+_77184416 | 0.77 |
ENST00000598970.1 |
DKFZP434E1119 |
DKFZP434E1119 |
| chr2_+_103089756 | 0.77 |
ENST00000295269.4 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr14_+_23012122 | 0.77 |
ENST00000390534.1 |
TRAJ3 |
T cell receptor alpha joining 3 |
| chr17_-_76123101 | 0.76 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
| chr4_-_159644507 | 0.76 |
ENST00000307720.3 |
PPID |
peptidylprolyl isomerase D |
| chr5_-_169694286 | 0.76 |
ENST00000521416.1 ENST00000520344.1 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr1_-_224517823 | 0.75 |
ENST00000469968.1 ENST00000436927.1 ENST00000469075.1 ENST00000488718.1 ENST00000482491.1 ENST00000340871.4 ENST00000492281.1 ENST00000361463.3 ENST00000391875.2 ENST00000461546.1 |
NVL |
nuclear VCP-like |
| chr15_+_75491213 | 0.75 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr20_+_43803517 | 0.75 |
ENST00000243924.3 |
PI3 |
peptidase inhibitor 3, skin-derived |
| chr17_-_30668887 | 0.73 |
ENST00000581747.1 ENST00000583334.1 ENST00000580558.1 |
C17orf75 |
chromosome 17 open reading frame 75 |
| chr14_+_77787227 | 0.73 |
ENST00000216465.5 ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr14_-_102553371 | 0.73 |
ENST00000553585.1 ENST00000216281.8 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr3_+_48264816 | 0.73 |
ENST00000296435.2 ENST00000576243.1 |
CAMP |
cathelicidin antimicrobial peptide |
| chr14_-_21493123 | 0.73 |
ENST00000556147.1 ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2 |
NDRG family member 2 |
| chr4_+_108910870 | 0.72 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr8_-_8318847 | 0.72 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr8_+_56685701 | 0.72 |
ENST00000260129.5 |
TGS1 |
trimethylguanosine synthase 1 |
| chr19_-_51523275 | 0.72 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chrX_+_64887512 | 0.72 |
ENST00000360270.5 |
MSN |
moesin |
| chr3_+_52245458 | 0.72 |
ENST00000459884.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr19_+_751122 | 0.71 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr11_-_47869865 | 0.71 |
ENST00000530326.1 ENST00000532747.1 |
NUP160 |
nucleoporin 160kDa |
| chr21_-_34144157 | 0.71 |
ENST00000331923.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr1_-_1711508 | 0.71 |
ENST00000378625.1 |
NADK |
NAD kinase |
| chrX_-_152989531 | 0.69 |
ENST00000458587.2 ENST00000416815.1 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr1_-_225616515 | 0.69 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr14_-_106725723 | 0.69 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr7_+_150434430 | 0.69 |
ENST00000358647.3 |
GIMAP5 |
GTPase, IMAP family member 5 |
| chr16_+_1730338 | 0.69 |
ENST00000566691.1 ENST00000382710.4 |
HN1L |
hematological and neurological expressed 1-like |
| chr4_+_108911036 | 0.69 |
ENST00000505878.1 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr6_+_32811885 | 0.68 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr15_-_89764929 | 0.68 |
ENST00000268125.5 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr2_-_191878874 | 0.68 |
ENST00000392322.3 ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr9_+_138391805 | 0.67 |
ENST00000371785.1 |
MRPS2 |
mitochondrial ribosomal protein S2 |
| chr14_-_107049312 | 0.67 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chrX_+_15767971 | 0.67 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr9_+_134065506 | 0.67 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr12_+_4647950 | 0.66 |
ENST00000321524.7 ENST00000543041.1 ENST00000228843.9 ENST00000352618.4 ENST00000544927.1 |
RAD51AP1 |
RAD51 associated protein 1 |
| chr5_+_134094461 | 0.66 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr13_-_95248511 | 0.66 |
ENST00000261296.5 |
TGDS |
TDP-glucose 4,6-dehydratase |
| chr2_+_231577532 | 0.66 |
ENST00000258418.5 |
CAB39 |
calcium binding protein 39 |
| chrX_-_152989798 | 0.65 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr4_+_114214125 | 0.64 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chr19_+_10362577 | 0.64 |
ENST00000592514.1 ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr19_+_10362882 | 0.63 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr9_-_77567743 | 0.63 |
ENST00000376854.5 |
C9orf40 |
chromosome 9 open reading frame 40 |
| chr11_-_134123142 | 0.62 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr5_+_156696362 | 0.62 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr11_-_125773085 | 0.62 |
ENST00000227474.3 ENST00000534158.1 ENST00000529801.1 |
PUS3 |
pseudouridylate synthase 3 |
| chr19_+_39421556 | 0.61 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr18_+_13611763 | 0.61 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr19_-_14628645 | 0.61 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_-_6445809 | 0.61 |
ENST00000377855.2 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr21_+_42733870 | 0.60 |
ENST00000330714.3 ENST00000436410.1 ENST00000435611.1 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
| chr10_+_75936444 | 0.60 |
ENST00000372734.3 ENST00000541550.1 |
ADK |
adenosine kinase |
| chr8_-_101321584 | 0.60 |
ENST00000523167.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chrX_-_7895755 | 0.60 |
ENST00000444736.1 ENST00000537427.1 ENST00000442940.1 |
PNPLA4 |
patatin-like phospholipase domain containing 4 |
| chrX_-_148669116 | 0.59 |
ENST00000243314.5 |
MAGEA9B |
melanoma antigen family A, 9B |
| chr5_+_173472607 | 0.59 |
ENST00000303177.3 ENST00000519867.1 |
NSG2 |
Neuron-specific protein family member 2 |
| chr9_+_139780942 | 0.58 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr2_+_58655461 | 0.58 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr7_+_148395959 | 0.58 |
ENST00000325222.4 |
CUL1 |
cullin 1 |
| chrX_+_77359671 | 0.57 |
ENST00000373316.4 |
PGK1 |
phosphoglycerate kinase 1 |
| chr6_-_100016527 | 0.57 |
ENST00000523985.1 ENST00000518714.1 ENST00000520371.1 |
CCNC |
cyclin C |
| chr1_-_22109682 | 0.57 |
ENST00000400301.1 ENST00000532737.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr1_-_10856694 | 0.57 |
ENST00000377022.3 ENST00000344008.5 |
CASZ1 |
castor zinc finger 1 |
| chr16_-_33647696 | 0.56 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chrX_+_77359726 | 0.56 |
ENST00000442431.1 |
PGK1 |
phosphoglycerate kinase 1 |
| chr6_+_31465849 | 0.55 |
ENST00000399150.3 |
MICB |
MHC class I polypeptide-related sequence B |
| chrX_-_11445856 | 0.55 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr11_-_47870019 | 0.55 |
ENST00000378460.2 |
NUP160 |
nucleoporin 160kDa |
| chr12_+_51632600 | 0.55 |
ENST00000549555.1 ENST00000439799.2 ENST00000425012.2 |
DAZAP2 |
DAZ associated protein 2 |
| chr6_+_10694900 | 0.54 |
ENST00000379568.3 |
PAK1IP1 |
PAK1 interacting protein 1 |
| chr9_+_140513438 | 0.53 |
ENST00000462484.1 ENST00000334856.6 ENST00000460843.1 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
| chr2_-_234763147 | 0.53 |
ENST00000411486.2 ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP |
Holliday junction recognition protein |
| chr10_-_62332357 | 0.53 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_+_130020932 | 0.52 |
ENST00000484324.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr6_-_84140757 | 0.52 |
ENST00000541327.1 ENST00000369705.3 ENST00000543031.1 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr10_-_115423792 | 0.52 |
ENST00000369360.3 ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP |
nebulin-related anchoring protein |
| chrX_+_148863584 | 0.52 |
ENST00000439010.2 ENST00000298974.5 ENST00000522429.1 ENST00000519822.1 |
MAGEA9 |
melanoma antigen family A, 9 |
| chr2_-_11272234 | 0.51 |
ENST00000590207.1 ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1 |
AC062028.1 |
| chr19_-_54876558 | 0.51 |
ENST00000391742.2 ENST00000434277.2 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.7 | 9.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.6 | 3.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.6 | 1.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 1.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.6 | 1.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 1.6 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.5 | 2.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 1.2 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.4 | 1.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.4 | 1.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.3 | 0.9 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 2.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 1.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 3.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 0.7 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 2.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 5.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 1.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 0.6 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 0.5 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.2 | 1.7 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 0.5 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 0.5 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 1.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 6.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.3 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 4.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.8 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.8 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.4 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 3.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.2 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.1 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 3.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 12.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.3 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 2.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.4 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.8 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.9 | 5.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 3.2 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.6 | 2.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.5 | 1.5 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.5 | 2.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.5 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 1.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 1.2 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.4 | 2.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 1.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.4 | 1.9 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.4 | 1.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.8 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 2.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 3.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 1.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.3 | 0.6 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.3 | 2.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 5.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.3 | 1.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 2.6 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 | 1.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.2 | 1.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 0.7 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 6.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.7 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.2 | 0.9 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 1.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.2 | 1.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.2 | 1.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 2.8 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 1.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 1.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 2.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 0.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.7 | GO:0050717 | regulation of interleukin-1 alpha secretion(GO:0050705) positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.5 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.2 | 0.6 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 1.5 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.6 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 1.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 1.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.4 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.5 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.9 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 5.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.6 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.4 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 2.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.4 | GO:0002697 | regulation of immune effector process(GO:0002697) |
| 0.1 | 0.7 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.4 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.1 | 6.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 10.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 1.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.6 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.4 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.1 | 0.5 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.1 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 3.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 1.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 0.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 1.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.8 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.4 | GO:0006778 | porphyrin-containing compound metabolic process(GO:0006778) porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) heme metabolic process(GO:0042168) pigment catabolic process(GO:0046149) cofactor catabolic process(GO:0051187) |
| 0.1 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 4.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 3.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 1.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 1.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 3.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 1.3 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.9 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 9.1 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.5 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.9 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.4 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 1.2 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.4 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.1 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0003284 | septum primum development(GO:0003284) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) regulation of AV node cell action potential(GO:0098904) regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.6 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.8 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.0 | GO:2000078 | columnar/cuboidal epithelial cell maturation(GO:0002069) glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 3.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 5.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 1.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.2 | 1.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 0.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 1.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.0 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.2 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 2.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 0.8 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 6.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 5.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 5.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 4.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0098644 | complex of collagen trimers(GO:0098644) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 3.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 7.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 3.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 7.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 3.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 3.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |