Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
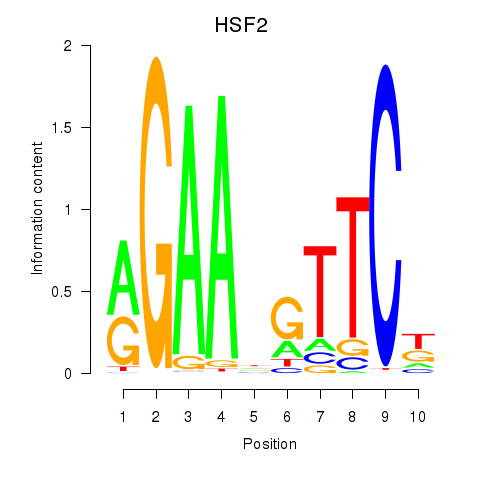
Results for HSF2
Z-value: 0.58
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | HSF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg19_v2_chr6_+_122720681_122720713 | 0.01 | 9.8e-01 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
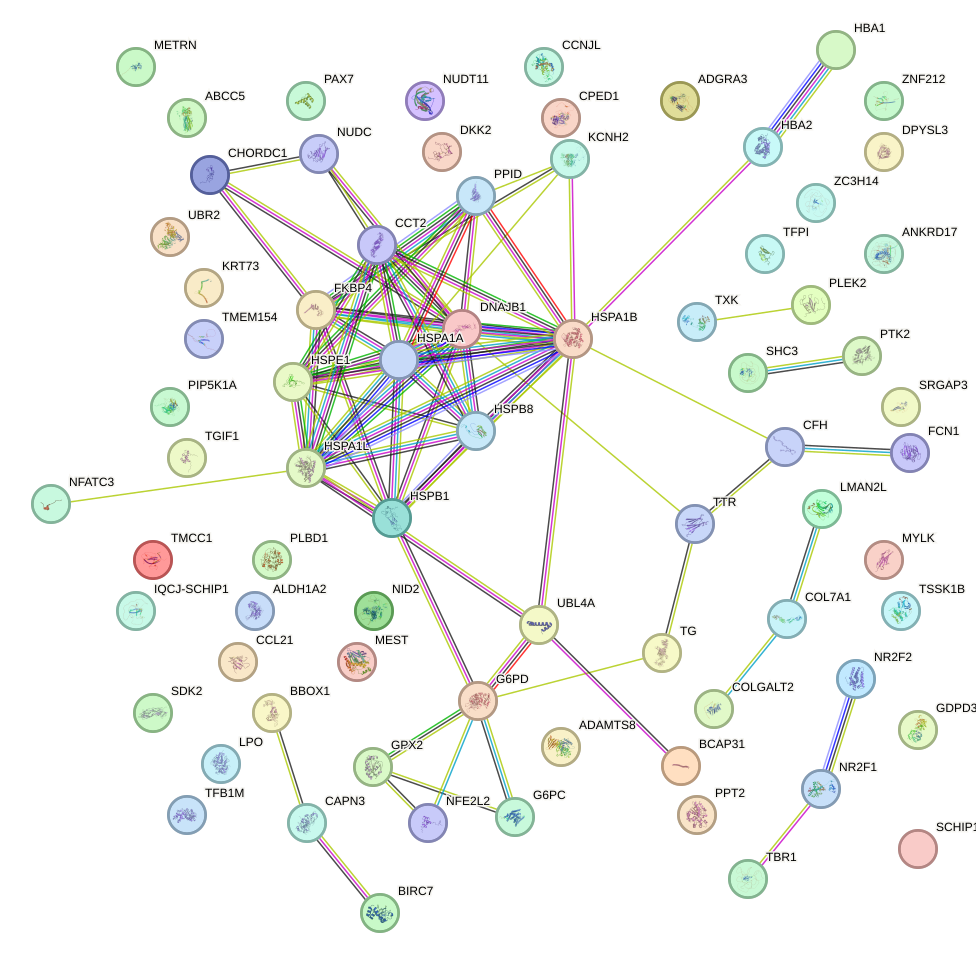
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_188419200 | 1.23 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_188419078 | 1.14 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr18_-_812231 | 1.01 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr6_+_31783291 | 0.92 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr2_-_219151487 | 0.91 |
ENST00000444881.1 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
| chr19_-_14629224 | 0.90 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr18_+_3449695 | 0.88 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr18_+_3449821 | 0.84 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr7_+_120629653 | 0.80 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr16_+_226658 | 0.77 |
ENST00000320868.5 ENST00000397797.1 |
HBA1 |
hemoglobin, alpha 1 |
| chr2_+_11674213 | 0.77 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr18_+_3451646 | 0.76 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr15_-_58357866 | 0.74 |
ENST00000537372.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr6_+_32121789 | 0.62 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chrX_-_102941596 | 0.61 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr6_+_151646800 | 0.60 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr18_+_3451584 | 0.60 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr4_-_22517620 | 0.59 |
ENST00000502482.1 ENST00000334304.5 |
GPR125 |
G protein-coupled receptor 125 |
| chr1_+_206516200 | 0.58 |
ENST00000295713.5 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr6_+_32121218 | 0.57 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr15_+_96869165 | 0.57 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_32121908 | 0.56 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_+_31795506 | 0.56 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr11_+_12696102 | 0.55 |
ENST00000527636.1 ENST00000527376.1 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr20_+_4666882 | 0.55 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr14_-_67859422 | 0.49 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr11_+_12695944 | 0.49 |
ENST00000361905.4 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr8_-_141931287 | 0.48 |
ENST00000517887.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr11_-_111781610 | 0.45 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr3_-_123411191 | 0.43 |
ENST00000354792.5 ENST00000508240.1 |
MYLK |
myosin light chain kinase |
| chrX_-_153775426 | 0.43 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr12_-_14721283 | 0.43 |
ENST00000240617.5 |
PLBD1 |
phospholipase B domain containing 1 |
| chr14_-_65409438 | 0.41 |
ENST00000557049.1 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr18_+_29171689 | 0.41 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr14_-_65409502 | 0.40 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr15_+_42651691 | 0.39 |
ENST00000357568.3 ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3 |
calpain 3, (p94) |
| chr9_-_34710066 | 0.39 |
ENST00000378792.1 ENST00000259607.2 |
CCL21 |
chemokine (C-C motif) ligand 21 |
| chr1_-_184006829 | 0.38 |
ENST00000361927.4 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chrX_+_135230712 | 0.37 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr7_-_150652924 | 0.35 |
ENST00000330883.4 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr14_-_52535712 | 0.34 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr12_-_53012343 | 0.33 |
ENST00000305748.3 |
KRT73 |
keratin 73 |
| chr11_-_111781454 | 0.32 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chrX_-_152989531 | 0.31 |
ENST00000458587.2 ENST00000416815.1 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr17_-_56494882 | 0.31 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494908 | 0.31 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chrX_-_51239425 | 0.30 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr17_-_56494713 | 0.30 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr11_+_75273101 | 0.28 |
ENST00000533603.1 ENST00000358171.3 ENST00000526242.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr16_+_765092 | 0.27 |
ENST00000568223.2 |
METRN |
meteorin, glial cell differentiation regulator |
| chrX_+_135278908 | 0.26 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr19_-_14628645 | 0.25 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_+_27062502 | 0.25 |
ENST00000263182.3 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_146833222 | 0.25 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_+_75273246 | 0.25 |
ENST00000526397.1 ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chrX_-_102942961 | 0.25 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chrX_-_102943022 | 0.24 |
ENST00000433176.2 |
MORF4L2 |
mortality factor 4 like 2 |
| chr3_-_183735731 | 0.24 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr4_-_186877806 | 0.24 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr16_+_68119764 | 0.23 |
ENST00000570212.1 ENST00000562926.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr4_-_153601136 | 0.23 |
ENST00000504064.1 ENST00000304385.3 |
TMEM154 |
transmembrane protein 154 |
| chr4_-_186877502 | 0.23 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chrX_+_135279179 | 0.23 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr11_+_27062860 | 0.22 |
ENST00000528583.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_146833485 | 0.22 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_-_111781554 | 0.21 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr2_-_178128250 | 0.21 |
ENST00000448782.1 ENST00000446151.2 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr6_+_149539767 | 0.20 |
ENST00000606202.1 ENST00000536230.1 ENST00000445901.1 |
TAB2 RP1-111D6.3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chrX_+_101478829 | 0.20 |
ENST00000372763.1 ENST00000372758.1 |
NXF2 |
nuclear RNA export factor 2 |
| chrX_-_101718085 | 0.19 |
ENST00000372750.1 ENST00000372752.1 |
NXF2B |
nuclear RNA export factor 2B |
| chr3_-_129407535 | 0.18 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr16_-_30125177 | 0.17 |
ENST00000406256.3 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_-_97405775 | 0.17 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr4_-_107957454 | 0.17 |
ENST00000285311.3 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
| chr11_-_89956227 | 0.17 |
ENST00000457199.2 ENST00000530765.1 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr1_+_27248203 | 0.17 |
ENST00000321265.5 |
NUDC |
nudC nuclear distribution protein |
| chr14_+_89060739 | 0.16 |
ENST00000318308.6 |
ZC3H14 |
zinc finger CCCH-type containing 14 |
| chr14_+_89060749 | 0.15 |
ENST00000555900.1 ENST00000406216.3 ENST00000557737.1 |
ZC3H14 |
zinc finger CCCH-type containing 14 |
| chr11_-_130298888 | 0.15 |
ENST00000257359.6 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr6_-_155635583 | 0.15 |
ENST00000367166.4 |
TFB1M |
transcription factor B1, mitochondrial |
| chr12_+_2904102 | 0.13 |
ENST00000001008.4 |
FKBP4 |
FK506 binding protein 4, 59kDa |
| chr17_+_41052808 | 0.13 |
ENST00000592383.1 ENST00000253801.2 ENST00000585489.1 |
G6PC |
glucose-6-phosphatase, catalytic subunit |
| chr7_+_148936732 | 0.13 |
ENST00000335870.2 |
ZNF212 |
zinc finger protein 212 |
| chr2_+_198365095 | 0.13 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr3_-_48632593 | 0.12 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr3_+_158787041 | 0.11 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr6_+_42584847 | 0.10 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr6_-_31782813 | 0.10 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr11_-_414948 | 0.10 |
ENST00000530494.1 ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chrX_-_72347916 | 0.09 |
ENST00000373518.1 |
NAP1L6 |
nucleosome assembly protein 1-like 6 |
| chr2_-_152589670 | 0.09 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr17_-_71410794 | 0.09 |
ENST00000424778.1 |
SDK2 |
sidekick cell adhesion molecule 2 |
| chr9_-_137809718 | 0.09 |
ENST00000371806.3 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
| chrX_+_153775821 | 0.09 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr4_-_74088800 | 0.09 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr7_-_36764142 | 0.08 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr7_+_75931861 | 0.08 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr1_+_151171012 | 0.08 |
ENST00000349792.5 ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr1_+_18958008 | 0.08 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr17_+_56315936 | 0.08 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr7_-_14880892 | 0.08 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr2_+_162272605 | 0.08 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr12_+_69979113 | 0.08 |
ENST00000299300.6 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr6_+_44215603 | 0.08 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr7_-_36764004 | 0.08 |
ENST00000431169.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr2_-_10978103 | 0.07 |
ENST00000404824.2 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr12_+_119616447 | 0.07 |
ENST00000281938.2 |
HSPB8 |
heat shock 22kDa protein 8 |
| chr4_-_48136217 | 0.07 |
ENST00000264316.4 |
TXK |
TXK tyrosine kinase |
| chr1_-_67142710 | 0.06 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr11_-_89956461 | 0.06 |
ENST00000320585.6 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr5_-_159739532 | 0.06 |
ENST00000520748.1 ENST00000393977.3 ENST00000257536.7 |
CCNJL |
cyclin J-like |
| chr3_-_49131013 | 0.06 |
ENST00000424300.1 |
QRICH1 |
glutamine-rich 1 |
| chr9_-_91793675 | 0.06 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr8_+_134125727 | 0.05 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr7_+_130126165 | 0.05 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr20_+_61867235 | 0.05 |
ENST00000342412.6 ENST00000217169.3 |
BIRC7 |
baculoviral IAP repeat containing 7 |
| chr5_-_112770674 | 0.05 |
ENST00000390666.3 |
TSSK1B |
testis-specific serine kinase 1B |
| chr4_-_159644507 | 0.05 |
ENST00000307720.3 |
PPID |
peptidylprolyl isomerase D |
| chr2_-_85581623 | 0.05 |
ENST00000449375.1 ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT |
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr16_+_71392616 | 0.04 |
ENST00000349553.5 ENST00000302628.4 ENST00000562305.1 |
CALB2 |
calbindin 2 |
| chr4_-_122085469 | 0.04 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr2_+_28615669 | 0.04 |
ENST00000379619.1 ENST00000264716.4 |
FOSL2 |
FOS-like antigen 2 |
| chr7_+_130126012 | 0.04 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr2_-_85581701 | 0.04 |
ENST00000295802.4 |
RETSAT |
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr12_+_69979210 | 0.04 |
ENST00000544368.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr5_+_79331164 | 0.03 |
ENST00000350881.2 |
THBS4 |
thrombospondin 4 |
| chr4_+_184427235 | 0.03 |
ENST00000412117.1 ENST00000434682.2 |
ING2 |
inhibitor of growth family, member 2 |
| chr10_+_135340859 | 0.03 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr16_-_30457048 | 0.03 |
ENST00000500504.2 ENST00000542752.1 |
SEPHS2 |
selenophosphate synthetase 2 |
| chr12_-_45269430 | 0.03 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr15_+_41952591 | 0.03 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr6_-_53013620 | 0.03 |
ENST00000259803.7 |
GCM1 |
glial cells missing homolog 1 (Drosophila) |
| chr12_-_45270077 | 0.03 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr6_+_153019023 | 0.02 |
ENST00000367245.5 ENST00000529453.1 |
MYCT1 |
myc target 1 |
| chr10_+_98064085 | 0.02 |
ENST00000419175.1 ENST00000371174.2 |
DNTT |
DNA nucleotidylexotransferase |
| chr12_+_69979446 | 0.02 |
ENST00000543146.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr5_-_135701164 | 0.02 |
ENST00000355180.3 ENST00000426057.2 ENST00000513104.1 |
TRPC7 |
transient receptor potential cation channel, subfamily C, member 7 |
| chr12_-_123011476 | 0.02 |
ENST00000528279.1 ENST00000344591.4 ENST00000526560.2 |
RSRC2 |
arginine/serine-rich coiled-coil 2 |
| chr6_+_146350267 | 0.01 |
ENST00000355289.4 ENST00000282753.1 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr20_+_55904815 | 0.01 |
ENST00000371263.3 ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11 |
SPO11 meiotic protein covalently bound to DSB |
| chr7_-_4901625 | 0.01 |
ENST00000404991.1 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
| chr19_+_17416609 | 0.01 |
ENST00000602206.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr17_+_73663402 | 0.01 |
ENST00000355423.3 |
SAP30BP |
SAP30 binding protein |
| chr17_-_39661849 | 0.01 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr11_-_71791518 | 0.00 |
ENST00000537217.1 ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr17_+_35294075 | 0.00 |
ENST00000254457.5 |
LHX1 |
LIM homeobox 1 |
| chr19_+_6135646 | 0.00 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chr6_-_135375986 | 0.00 |
ENST00000525067.1 ENST00000367822.5 ENST00000367837.5 |
HBS1L |
HBS1-like (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.1 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 3.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 1.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.3 | 2.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 1.7 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.5 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.3 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 1.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.5 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.6 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.1 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 1.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |