Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
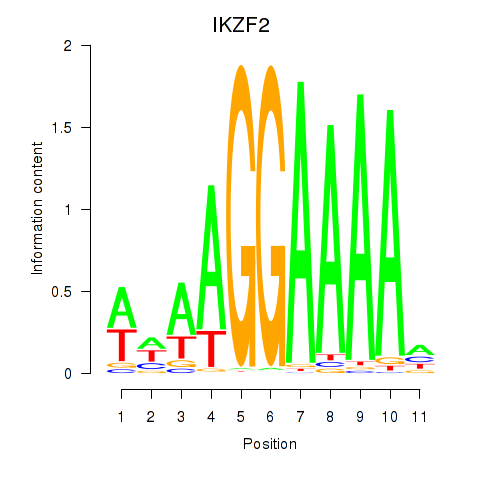
Results for IKZF2
Z-value: 1.22
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKZF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214014959_214015058 | 0.14 | 6.1e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 5.03 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr12_-_91573316 | 3.55 |
ENST00000393155.1 |
DCN |
decorin |
| chr1_-_57045228 | 3.51 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr2_-_190044480 | 3.37 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr13_-_38172863 | 3.22 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr12_-_91573132 | 3.17 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573249 | 3.14 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr7_+_93551011 | 2.39 |
ENST00000248564.5 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
| chr1_-_153518270 | 2.35 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr1_+_101185290 | 2.23 |
ENST00000370119.4 ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1 |
vascular cell adhesion molecule 1 |
| chr19_-_43709703 | 2.19 |
ENST00000599391.1 ENST00000244295.9 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr3_-_114035026 | 2.06 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr21_-_28338732 | 1.76 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr5_-_42812143 | 1.72 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr5_-_42811986 | 1.67 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr1_+_158979680 | 1.64 |
ENST00000368131.4 ENST00000340979.6 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 1.61 |
ENST00000368132.3 ENST00000295809.7 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chrX_+_135230712 | 1.35 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr5_-_111091948 | 1.32 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr1_-_203151933 | 1.31 |
ENST00000404436.2 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr12_+_26348246 | 1.30 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr8_+_97597148 | 1.28 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr1_+_158979792 | 1.25 |
ENST00000359709.3 ENST00000430894.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr12_+_26348429 | 1.21 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chrX_+_135251783 | 1.18 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chrX_+_100805496 | 1.16 |
ENST00000372829.3 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
| chr19_-_43709817 | 1.14 |
ENST00000433626.2 ENST00000405312.3 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chrX_+_135251835 | 1.08 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr4_-_71532339 | 1.06 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr18_-_53070913 | 1.02 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr19_-_43709772 | 1.01 |
ENST00000596907.1 ENST00000451895.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr4_-_157892498 | 1.01 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr4_+_26322185 | 1.00 |
ENST00000361572.6 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_104443890 | 0.99 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr6_-_152639479 | 0.98 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_+_32711419 | 0.97 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr6_-_131384347 | 0.97 |
ENST00000530481.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr6_-_131384412 | 0.96 |
ENST00000445890.2 ENST00000368128.2 ENST00000337057.3 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr15_-_90358048 | 0.95 |
ENST00000300060.6 ENST00000560137.1 |
ANPEP |
alanyl (membrane) aminopeptidase |
| chr6_-_131384373 | 0.93 |
ENST00000392427.3 ENST00000525271.1 ENST00000527411.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr2_-_190927447 | 0.93 |
ENST00000260950.4 |
MSTN |
myostatin |
| chrX_+_135252050 | 0.91 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr4_+_26322409 | 0.90 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_-_79398250 | 0.88 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_+_55478538 | 0.88 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr4_-_114900831 | 0.88 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr17_-_46688334 | 0.75 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr14_-_106926724 | 0.68 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr14_+_45431379 | 0.67 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr1_+_79115503 | 0.67 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr1_+_21880560 | 0.66 |
ENST00000425315.2 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr12_-_50677255 | 0.65 |
ENST00000551691.1 ENST00000394943.3 ENST00000341247.4 |
LIMA1 |
LIM domain and actin binding 1 |
| chr9_+_92219919 | 0.63 |
ENST00000252506.6 ENST00000375769.1 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
| chrX_+_80457442 | 0.63 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr15_-_20193370 | 0.63 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr12_+_26348582 | 0.61 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr19_-_43269809 | 0.59 |
ENST00000406636.3 ENST00000404209.4 ENST00000306511.4 |
PSG8 |
pregnancy specific beta-1-glycoprotein 8 |
| chr16_-_33647696 | 0.58 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr6_+_32605195 | 0.58 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr10_-_127505167 | 0.58 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr2_+_46926048 | 0.57 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr11_+_71903169 | 0.57 |
ENST00000393676.3 |
FOLR1 |
folate receptor 1 (adult) |
| chr5_+_32710736 | 0.56 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr7_+_73442487 | 0.56 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr6_+_32605134 | 0.56 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr9_+_34989638 | 0.55 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr14_+_64970662 | 0.54 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr2_+_46926326 | 0.54 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr14_-_106552755 | 0.52 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr8_-_18541603 | 0.52 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr8_+_26240414 | 0.51 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr13_-_99959641 | 0.51 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr8_-_17942432 | 0.50 |
ENST00000381733.4 ENST00000314146.10 |
ASAH1 |
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr4_+_72052964 | 0.49 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_+_124320665 | 0.48 |
ENST00000394339.2 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr18_-_21852143 | 0.47 |
ENST00000399443.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr12_-_6580094 | 0.47 |
ENST00000361716.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr6_+_123110465 | 0.47 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr9_-_36401155 | 0.47 |
ENST00000377885.2 |
RNF38 |
ring finger protein 38 |
| chr18_-_53089723 | 0.46 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr6_+_123110302 | 0.46 |
ENST00000368440.4 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr12_+_5541267 | 0.46 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr19_-_43244610 | 0.46 |
ENST00000595140.1 ENST00000327495.5 |
PSG3 |
pregnancy specific beta-1-glycoprotein 3 |
| chr17_-_10421853 | 0.46 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr1_-_68698222 | 0.44 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr7_+_73442457 | 0.43 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr10_-_112678976 | 0.43 |
ENST00000448814.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr19_-_43421989 | 0.41 |
ENST00000292125.2 ENST00000187910.2 |
PSG6 |
pregnancy specific beta-1-glycoprotein 6 |
| chr22_-_36220420 | 0.41 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr9_+_124062071 | 0.40 |
ENST00000373818.4 |
GSN |
gelsolin |
| chr5_+_43602750 | 0.39 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr12_-_80328949 | 0.39 |
ENST00000450142.2 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
| chr1_+_17944832 | 0.38 |
ENST00000167825.4 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr3_+_12392971 | 0.38 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr18_-_47340297 | 0.37 |
ENST00000586485.1 ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2 |
acetyl-CoA acyltransferase 2 |
| chr1_+_53480598 | 0.34 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr4_-_186733363 | 0.34 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_-_133706695 | 0.34 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr10_-_104192405 | 0.34 |
ENST00000369937.4 |
CUEDC2 |
CUE domain containing 2 |
| chr12_-_118797475 | 0.32 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr2_+_27301435 | 0.32 |
ENST00000380320.4 |
EMILIN1 |
elastin microfibril interfacer 1 |
| chr5_+_92919043 | 0.31 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr7_+_79765071 | 0.31 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr15_+_81475047 | 0.31 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr10_+_112679301 | 0.31 |
ENST00000265277.5 ENST00000369452.4 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
| chr5_-_96518907 | 0.31 |
ENST00000508447.1 ENST00000283109.3 |
RIOK2 |
RIO kinase 2 |
| chr1_+_104615595 | 0.31 |
ENST00000418362.1 |
RP11-364B6.1 |
RP11-364B6.1 |
| chr10_-_112678904 | 0.31 |
ENST00000423273.1 ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr19_-_43422019 | 0.30 |
ENST00000402603.4 ENST00000594375.1 |
PSG6 |
pregnancy specific beta-1-glycoprotein 6 |
| chr19_+_10197463 | 0.30 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr1_+_158801095 | 0.30 |
ENST00000368141.4 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr2_+_33661382 | 0.29 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr14_-_106668095 | 0.29 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr15_-_63449663 | 0.29 |
ENST00000439025.1 |
RPS27L |
ribosomal protein S27-like |
| chr7_+_73442102 | 0.29 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chrX_-_135338503 | 0.28 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chrX_+_49832231 | 0.28 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr14_-_107114267 | 0.28 |
ENST00000454421.2 |
IGHV3-64 |
immunoglobulin heavy variable 3-64 |
| chr9_-_74980113 | 0.27 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr10_-_98031265 | 0.27 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr8_-_19459993 | 0.27 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_+_39184024 | 0.27 |
ENST00000399820.3 ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19 |
WD repeat domain 19 |
| chr1_+_209941942 | 0.27 |
ENST00000487271.1 ENST00000477431.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr10_-_112678692 | 0.27 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chrX_-_43741594 | 0.26 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr4_+_68424434 | 0.26 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr20_-_3996036 | 0.25 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chrX_+_78003204 | 0.25 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr10_+_13628933 | 0.24 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr4_-_186696425 | 0.24 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_-_28113965 | 0.24 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chr9_+_2159850 | 0.24 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_-_44031375 | 0.23 |
ENST00000292147.2 |
ETHE1 |
ethylmalonic encephalopathy 1 |
| chrX_+_73164149 | 0.23 |
ENST00000602938.1 ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX |
JPX transcript, XIST activator (non-protein coding) |
| chr2_-_45795145 | 0.23 |
ENST00000535761.1 |
SRBD1 |
S1 RNA binding domain 1 |
| chr9_-_6015607 | 0.22 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr19_-_44031341 | 0.22 |
ENST00000600651.1 |
ETHE1 |
ethylmalonic encephalopathy 1 |
| chr9_+_19049372 | 0.22 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr15_-_59665062 | 0.20 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr5_+_43603229 | 0.20 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr7_+_120628731 | 0.19 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_74221288 | 0.19 |
ENST00000451013.2 |
GTF2IRD2 |
GTF2I repeat domain containing 2 |
| chr13_+_20532848 | 0.19 |
ENST00000382874.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr13_-_73356009 | 0.19 |
ENST00000377780.4 ENST00000377767.4 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
| chr4_-_83765613 | 0.18 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr8_+_30244580 | 0.18 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr6_+_26365443 | 0.18 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr4_-_74486217 | 0.18 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chrX_+_100663243 | 0.18 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr2_-_33824336 | 0.18 |
ENST00000431950.1 ENST00000403368.1 ENST00000441530.2 |
FAM98A |
family with sequence similarity 98, member A |
| chr11_+_11863579 | 0.18 |
ENST00000399455.2 |
USP47 |
ubiquitin specific peptidase 47 |
| chr3_+_10068095 | 0.17 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr10_-_98031310 | 0.17 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr10_-_76868931 | 0.17 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr13_+_20532900 | 0.16 |
ENST00000382871.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr3_-_52931557 | 0.16 |
ENST00000504329.1 ENST00000355083.5 |
TMEM110-MUSTN1 TMEM110 |
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr17_-_16118835 | 0.16 |
ENST00000582357.1 ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1 |
nuclear receptor corepressor 1 |
| chr3_-_18466026 | 0.15 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr11_-_72385437 | 0.15 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr14_-_92506371 | 0.15 |
ENST00000267622.4 |
TRIP11 |
thyroid hormone receptor interactor 11 |
| chr8_-_101964231 | 0.15 |
ENST00000521309.1 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_-_74486347 | 0.14 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_-_108093329 | 0.14 |
ENST00000278612.8 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
| chr17_+_7792101 | 0.14 |
ENST00000358181.4 ENST00000330494.7 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr19_-_12777509 | 0.14 |
ENST00000221363.4 ENST00000598876.1 ENST00000456935.2 ENST00000486847.2 |
MAN2B1 |
mannosidase, alpha, class 2B, member 1 |
| chr4_+_74347400 | 0.14 |
ENST00000226355.3 |
AFM |
afamin |
| chr12_-_49449107 | 0.14 |
ENST00000301067.7 |
KMT2D |
lysine (K)-specific methyltransferase 2D |
| chr13_-_46756351 | 0.13 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_-_151574191 | 0.13 |
ENST00000287878.4 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr3_-_18480260 | 0.13 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr17_-_73127778 | 0.12 |
ENST00000578407.1 |
NT5C |
5', 3'-nucleotidase, cytosolic |
| chr17_-_39197699 | 0.12 |
ENST00000306271.4 |
KRTAP1-1 |
keratin associated protein 1-1 |
| chr9_-_6470375 | 0.12 |
ENST00000355513.4 |
C9orf38 |
chromosome 9 open reading frame 38 |
| chr3_+_178525080 | 0.12 |
ENST00000358316.3 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr2_-_174830430 | 0.11 |
ENST00000310015.6 ENST00000455789.2 |
SP3 |
Sp3 transcription factor |
| chr8_+_85095553 | 0.11 |
ENST00000521268.1 |
RALYL |
RALY RNA binding protein-like |
| chr6_-_18265050 | 0.11 |
ENST00000397239.3 |
DEK |
DEK oncogene |
| chr10_+_105036909 | 0.11 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr7_-_135194822 | 0.11 |
ENST00000428680.2 ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4 |
CCR4-NOT transcription complex, subunit 4 |
| chr1_-_48866517 | 0.11 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr1_-_155948318 | 0.11 |
ENST00000361247.4 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_44220162 | 0.10 |
ENST00000244314.5 |
IRGC |
immunity-related GTPase family, cinema |
| chr22_-_50699701 | 0.10 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr13_-_48669232 | 0.10 |
ENST00000258648.2 ENST00000378586.1 |
MED4 |
mediator complex subunit 4 |
| chr3_+_69812877 | 0.10 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_143266297 | 0.10 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr12_+_53835508 | 0.09 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr4_-_89978299 | 0.09 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr16_+_81272287 | 0.09 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr11_-_65430251 | 0.09 |
ENST00000534283.1 ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA |
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr5_+_122847781 | 0.09 |
ENST00000395412.1 ENST00000395411.1 ENST00000345990.4 |
CSNK1G3 |
casein kinase 1, gamma 3 |
| chr12_+_62860581 | 0.08 |
ENST00000393632.2 ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2 |
MON2 homolog (S. cerevisiae) |
| chr2_-_136288113 | 0.08 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr1_+_10490127 | 0.08 |
ENST00000602787.1 ENST00000602296.1 ENST00000400900.2 |
APITD1 APITD1-CORT |
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr9_-_131790464 | 0.08 |
ENST00000417224.1 ENST00000416629.1 ENST00000372559.1 |
SH3GLB2 |
SH3-domain GRB2-like endophilin B2 |
| chr1_-_15850676 | 0.08 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr3_+_130650738 | 0.08 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_+_44220247 | 0.08 |
ENST00000596627.1 |
IRGC |
immunity-related GTPase family, cinema |
| chr12_+_60058458 | 0.07 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_190722119 | 0.06 |
ENST00000452382.1 |
PMS1 |
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.7 | 9.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 5.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 2.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 3.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0043596 | replication fork(GO:0005657) nuclear replication fork(GO:0043596) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 8.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.7 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 3.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 2.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 1.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 11.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.3 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 2.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.5 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 3.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 9.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 4.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.8 | 3.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.8 | 9.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.6 | 1.9 | GO:1905066 | arterial endothelial cell fate commitment(GO:0060844) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.6 | 5.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 2.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.3 | 0.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 3.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.7 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 0.6 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.2 | 1.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.5 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.6 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 1.0 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 4.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.2 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 4.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.4 | GO:1903921 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) protein processing in phagocytic vesicle(GO:1900756) regulation of plasma membrane raft polarization(GO:1903906) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.5 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.4 | GO:1902109 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.3 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 2.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.3 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 1.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.6 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |