Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
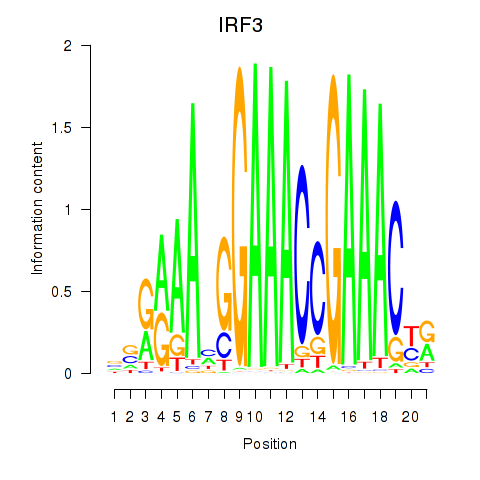
Results for IRF3
Z-value: 1.33
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | IRF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF3 | hg19_v2_chr19_-_50169064_50169132 | 0.12 | 6.6e-01 | Click! |
Activity profile of IRF3 motif
Sorted Z-values of IRF3 motif
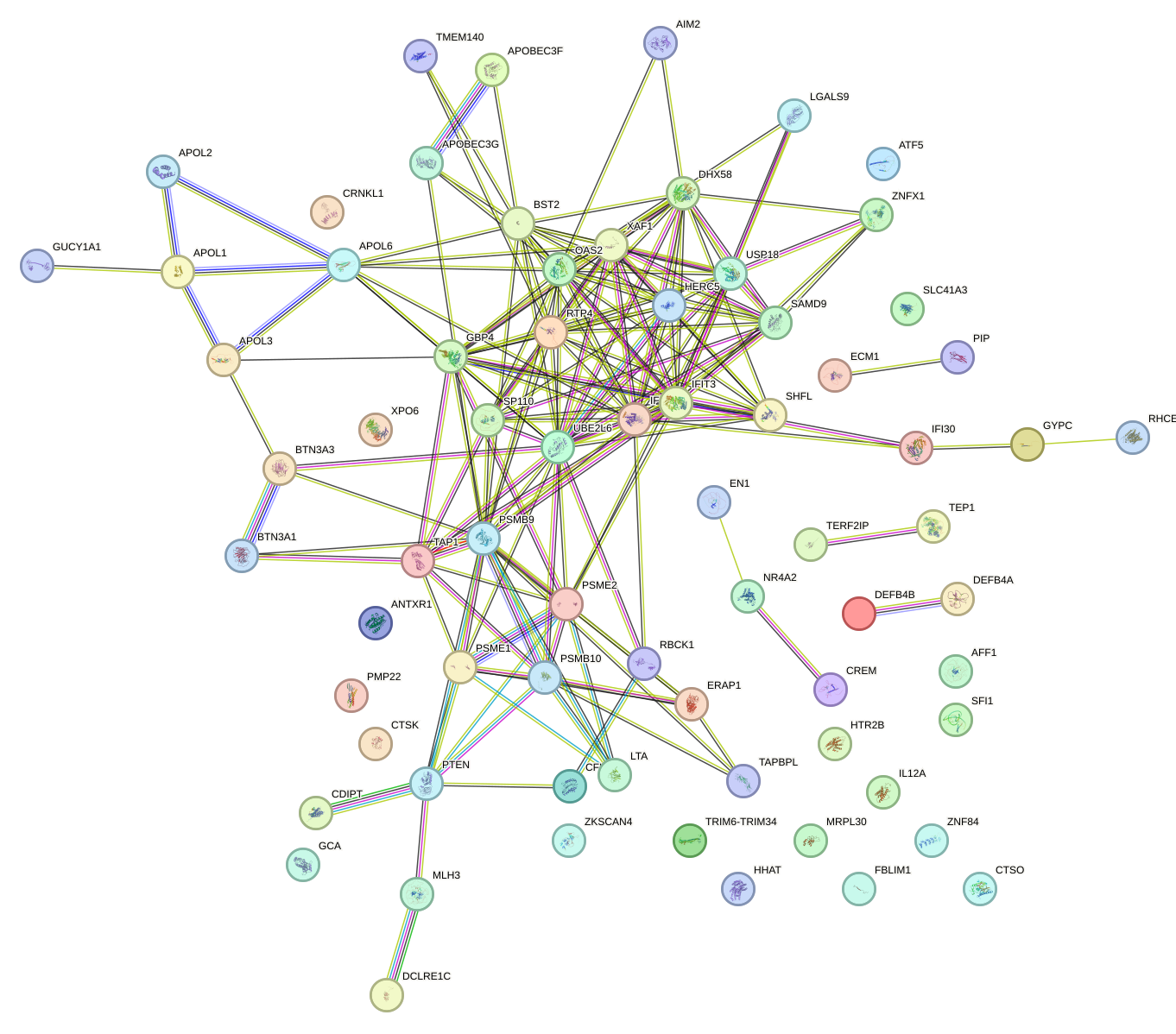
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_127413677 | 4.22 |
ENST00000356887.7 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr2_+_127413704 | 4.20 |
ENST00000409836.3 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr2_+_127413481 | 4.14 |
ENST00000259254.4 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr5_-_95158644 | 2.94 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr1_+_948803 | 2.77 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr19_+_18284477 | 2.48 |
ENST00000407280.3 |
IFI30 |
interferon, gamma-inducible protein 30 |
| chr6_+_32821924 | 2.08 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_-_95158375 | 2.01 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr21_+_42792442 | 2.00 |
ENST00000398600.2 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_-_57335280 | 1.94 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr6_+_31540056 | 1.39 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr1_-_159046617 | 1.32 |
ENST00000368130.4 |
AIM2 |
absent in melanoma 2 |
| chr16_-_67970990 | 1.25 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr16_-_11350036 | 1.22 |
ENST00000332029.2 |
SOCS1 |
suppressor of cytokine signaling 1 |
| chr4_+_89378261 | 1.18 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr1_+_79115503 | 1.06 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr19_-_17516449 | 0.96 |
ENST00000252593.6 |
BST2 |
bone marrow stromal cell antigen 2 |
| chr19_+_50431959 | 0.96 |
ENST00000595125.1 |
ATF5 |
activating transcription factor 5 |
| chr10_+_91087651 | 0.95 |
ENST00000371818.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_-_231084617 | 0.92 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr14_+_24605361 | 0.90 |
ENST00000206451.6 ENST00000559123.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_-_163175133 | 0.88 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr14_+_24605389 | 0.86 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_+_50432400 | 0.86 |
ENST00000423777.2 ENST00000600336.1 ENST00000597227.1 |
ATF5 |
activating transcription factor 5 |
| chr17_+_6659153 | 0.83 |
ENST00000441631.1 ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1 |
XIAP associated factor 1 |
| chr4_-_156875003 | 0.77 |
ENST00000433477.3 |
CTSO |
cathepsin O |
| chr22_+_18632666 | 0.75 |
ENST00000215794.7 |
USP18 |
ubiquitin specific peptidase 18 |
| chr1_+_76190357 | 0.73 |
ENST00000370834.5 ENST00000541113.1 ENST00000543667.1 ENST00000420607.2 |
ACADM |
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr10_+_91061712 | 0.73 |
ENST00000371826.3 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
| chr17_-_40264692 | 0.68 |
ENST00000591220.1 ENST00000251642.3 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr6_-_28226984 | 0.68 |
ENST00000423974.2 |
ZKSCAN4 |
zinc finger with KRAB and SCAN domains 4 |
| chr16_+_75681650 | 0.66 |
ENST00000300086.4 |
TERF2IP |
telomeric repeat binding factor 2, interacting protein |
| chr10_+_35415978 | 0.66 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr2_-_231084820 | 0.63 |
ENST00000258382.5 ENST00000338556.3 |
SP110 |
SP110 nuclear body protein |
| chr2_+_201981527 | 0.63 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_-_231084659 | 0.62 |
ENST00000258381.6 ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110 |
SP110 nuclear body protein |
| chr6_+_26440700 | 0.61 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr6_-_32821599 | 0.59 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr15_-_55611306 | 0.59 |
ENST00000563262.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_+_113416191 | 0.54 |
ENST00000342315.4 ENST00000392583.2 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr11_-_615570 | 0.52 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr17_+_36861735 | 0.52 |
ENST00000378137.5 ENST00000325718.7 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr22_+_36649056 | 0.50 |
ENST00000397278.3 ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1 |
apolipoprotein L, 1 |
| chr22_+_36044411 | 0.48 |
ENST00000409652.4 |
APOL6 |
apolipoprotein L, 6 |
| chr3_+_187086120 | 0.46 |
ENST00000259030.2 |
RTP4 |
receptor (chemosensory) transporter protein 4 |
| chr2_-_157198860 | 0.45 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_69240302 | 0.45 |
ENST00000303714.4 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr1_+_25598989 | 0.44 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chr2_+_69240415 | 0.43 |
ENST00000409829.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr3_-_53079281 | 0.43 |
ENST00000394750.1 |
SFMBT1 |
Scm-like with four mbt domains 1 |
| chr20_-_20033052 | 0.42 |
ENST00000536226.1 |
CRNKL1 |
crooked neck pre-mRNA splicing factor 1 |
| chr17_+_25958174 | 0.41 |
ENST00000313648.6 ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
| chr2_-_145275228 | 0.41 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr11_-_615942 | 0.41 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr19_+_10196781 | 0.40 |
ENST00000253110.11 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr2_+_163175394 | 0.40 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr2_+_99771418 | 0.39 |
ENST00000393473.2 ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1 MRPL30 |
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr16_-_29874211 | 0.38 |
ENST00000563415.1 |
CDIPT |
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr4_+_156588350 | 0.38 |
ENST00000296518.7 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr2_-_231989808 | 0.38 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_156588249 | 0.38 |
ENST00000393832.3 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr5_-_96143602 | 0.38 |
ENST00000443439.2 ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr11_+_5646213 | 0.36 |
ENST00000429814.2 |
TRIM34 |
tripartite motif containing 34 |
| chr3_+_159706537 | 0.35 |
ENST00000305579.2 ENST00000480787.1 ENST00000466512.1 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr14_-_75518129 | 0.35 |
ENST00000556257.1 ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3 |
mutL homolog 3 |
| chr10_+_35416223 | 0.35 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr12_+_6561190 | 0.35 |
ENST00000544021.1 ENST00000266556.7 |
TAPBPL |
TAP binding protein-like |
| chr20_-_47894936 | 0.35 |
ENST00000371754.4 |
ZNFX1 |
zinc finger, NFX1-type containing 1 |
| chr7_-_92747269 | 0.34 |
ENST00000446617.1 ENST00000379958.2 |
SAMD9 |
sterile alpha motif domain containing 9 |
| chr8_-_7274385 | 0.34 |
ENST00000318157.2 |
DEFB4B |
defensin, beta 4B |
| chr6_+_26365443 | 0.34 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr14_-_24615805 | 0.34 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr2_+_201981119 | 0.34 |
ENST00000395148.2 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr14_-_24615523 | 0.33 |
ENST00000559056.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr22_-_36556821 | 0.33 |
ENST00000531095.1 ENST00000397293.2 ENST00000349314.2 |
APOL3 |
apolipoprotein L, 3 |
| chr2_-_220252603 | 0.32 |
ENST00000322176.7 ENST00000273075.4 |
DNPEP |
aspartyl aminopeptidase |
| chr15_-_62352570 | 0.31 |
ENST00000261517.5 ENST00000395896.4 ENST00000395898.3 |
VPS13C |
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr19_+_50887585 | 0.31 |
ENST00000440232.2 ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1 |
polymerase (DNA directed), delta 1, catalytic subunit |
| chr4_+_100495864 | 0.30 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr7_+_134832808 | 0.30 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr10_-_14996017 | 0.30 |
ENST00000378241.1 ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr4_+_156588115 | 0.29 |
ENST00000455639.2 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr1_+_210502238 | 0.29 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr14_-_91976794 | 0.28 |
ENST00000555462.1 |
SMEK1 |
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr20_+_388791 | 0.27 |
ENST00000441733.1 ENST00000353660.3 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr6_-_41039567 | 0.27 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr10_-_70231639 | 0.25 |
ENST00000551118.2 ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2 |
DNA replication helicase/nuclease 2 |
| chr8_+_145438870 | 0.24 |
ENST00000527931.1 |
FAM203B |
family with sequence similarity 203, member B |
| chr6_+_26402465 | 0.23 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr1_-_89664595 | 0.22 |
ENST00000355754.6 |
GBP4 |
guanylate binding protein 4 |
| chr16_-_28223166 | 0.21 |
ENST00000304658.5 |
XPO6 |
exportin 6 |
| chr1_-_25747283 | 0.21 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr1_+_25598872 | 0.21 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr4_+_87856129 | 0.20 |
ENST00000395146.4 ENST00000507468.1 |
AFF1 |
AF4/FMR2 family, member 1 |
| chr10_-_14996070 | 0.20 |
ENST00000378258.1 ENST00000453695.2 ENST00000378246.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr7_+_142829162 | 0.20 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr22_+_31892261 | 0.19 |
ENST00000432498.1 ENST00000540643.1 ENST00000443326.1 ENST00000414585.1 |
SFI1 |
Sfi1 homolog, spindle assembly associated (yeast) |
| chr15_+_64428529 | 0.19 |
ENST00000560861.1 |
SNX1 |
sorting nexin 1 |
| chr22_+_31892373 | 0.19 |
ENST00000443011.1 ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1 |
Sfi1 homolog, spindle assembly associated (yeast) |
| chr3_-_125803105 | 0.18 |
ENST00000346785.5 ENST00000315891.6 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr19_+_49977466 | 0.18 |
ENST00000596435.1 ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr5_-_96143796 | 0.16 |
ENST00000296754.3 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr12_+_133614119 | 0.16 |
ENST00000327668.7 |
ZNF84 |
zinc finger protein 84 |
| chr10_+_91092241 | 0.15 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr14_-_20881579 | 0.15 |
ENST00000556935.1 ENST00000262715.5 ENST00000556549.1 |
TEP1 |
telomerase-associated protein 1 |
| chr2_-_119605253 | 0.15 |
ENST00000295206.6 |
EN1 |
engrailed homeobox 1 |
| chr4_+_156587853 | 0.15 |
ENST00000506455.1 ENST00000511108.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr3_-_125802765 | 0.14 |
ENST00000514891.1 ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr4_+_156587979 | 0.14 |
ENST00000511507.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr1_+_25599018 | 0.14 |
ENST00000417538.2 ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD |
Rh blood group, D antigen |
| chr16_-_29874462 | 0.13 |
ENST00000566113.1 ENST00000569956.1 ENST00000570016.1 |
CDIPT |
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr1_+_150480576 | 0.13 |
ENST00000346569.6 |
ECM1 |
extracellular matrix protein 1 |
| chr1_+_150480551 | 0.13 |
ENST00000369049.4 ENST00000369047.4 |
ECM1 |
extracellular matrix protein 1 |
| chr22_+_39436862 | 0.13 |
ENST00000381565.2 ENST00000452957.2 |
APOBEC3F APOBEC3G |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr12_+_12224331 | 0.12 |
ENST00000396367.1 ENST00000266434.4 ENST00000396369.1 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr8_+_7752151 | 0.12 |
ENST00000302247.2 |
DEFB4A |
defensin, beta 4A |
| chr12_-_322821 | 0.12 |
ENST00000359674.4 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr20_+_388679 | 0.12 |
ENST00000356286.5 ENST00000475269.1 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr17_+_21729899 | 0.12 |
ENST00000583708.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr9_-_95087604 | 0.12 |
ENST00000542613.1 ENST00000542053.1 ENST00000358855.4 ENST00000545558.1 ENST00000432670.2 ENST00000433029.2 ENST00000411621.2 |
NOL8 |
nucleolar protein 8 |
| chr6_+_134758827 | 0.11 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr17_+_21730180 | 0.11 |
ENST00000584398.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr4_+_118955500 | 0.11 |
ENST00000296499.5 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_+_133613878 | 0.11 |
ENST00000392319.2 ENST00000543758.1 |
ZNF84 |
zinc finger protein 84 |
| chr9_-_135282195 | 0.11 |
ENST00000334270.2 |
TTF1 |
transcription termination factor, RNA polymerase I |
| chr14_+_73525229 | 0.10 |
ENST00000527432.1 ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25 |
RNA binding motif protein 25 |
| chr11_+_118230287 | 0.10 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chr6_+_28193037 | 0.10 |
ENST00000531981.1 ENST00000425468.2 ENST00000252207.5 ENST00000531979.1 ENST00000527436.1 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
| chr6_-_36355513 | 0.10 |
ENST00000340181.4 ENST00000373737.4 |
ETV7 |
ets variant 7 |
| chr12_-_54582655 | 0.09 |
ENST00000504338.1 ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1 |
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr12_-_48963829 | 0.09 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr4_+_87928140 | 0.09 |
ENST00000307808.6 |
AFF1 |
AF4/FMR2 family, member 1 |
| chr14_+_77790901 | 0.09 |
ENST00000553586.1 ENST00000555583.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr14_+_73525265 | 0.09 |
ENST00000525161.1 |
RBM25 |
RNA binding motif protein 25 |
| chr14_+_73525144 | 0.09 |
ENST00000261973.7 ENST00000540173.1 |
RBM25 |
RNA binding motif protein 25 |
| chr9_-_95087838 | 0.09 |
ENST00000442668.2 ENST00000421075.2 ENST00000536624.1 |
NOL8 |
nucleolar protein 8 |
| chrX_+_77154935 | 0.09 |
ENST00000481445.1 |
COX7B |
cytochrome c oxidase subunit VIIb |
| chr1_-_146644122 | 0.08 |
ENST00000254101.3 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr5_-_77656175 | 0.07 |
ENST00000513755.1 ENST00000421004.3 |
CTD-2037K23.2 |
CTD-2037K23.2 |
| chr16_-_29875057 | 0.07 |
ENST00000219789.6 |
CDIPT |
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr17_-_30669138 | 0.06 |
ENST00000225805.4 ENST00000577809.1 |
C17orf75 |
chromosome 17 open reading frame 75 |
| chr12_+_133613937 | 0.05 |
ENST00000539354.1 ENST00000542874.1 ENST00000438628.2 |
ZNF84 |
zinc finger protein 84 |
| chr7_-_138794081 | 0.05 |
ENST00000464606.1 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
| chr2_-_37384175 | 0.05 |
ENST00000411537.2 ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2 |
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr4_-_74847800 | 0.05 |
ENST00000296029.3 |
PF4 |
platelet factor 4 |
| chr16_-_75681522 | 0.05 |
ENST00000568378.1 ENST00000568682.1 ENST00000570215.1 ENST00000319410.5 ENST00000302445.3 |
KARS |
lysyl-tRNA synthetase |
| chr17_-_63052929 | 0.03 |
ENST00000439174.2 |
GNA13 |
guanine nucleotide binding protein (G protein), alpha 13 |
| chr1_-_146644036 | 0.03 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr12_+_12223867 | 0.02 |
ENST00000308721.5 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr12_+_4758264 | 0.02 |
ENST00000266544.5 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr2_-_175629135 | 0.01 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr19_+_10196981 | 0.01 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr20_+_388935 | 0.00 |
ENST00000382181.2 ENST00000400247.3 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_+_151129135 | 0.00 |
ENST00000602841.1 |
SCNM1 |
sodium channel modifier 1 |
| chr15_+_55611128 | 0.00 |
ENST00000164305.5 ENST00000539642.1 |
PIGB |
phosphatidylinositol glycan anchor biosynthesis, class B |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 5.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.3 | 2.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 2.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 0.6 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.5 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 3.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.4 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.5 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.4 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 11.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 1.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.4 | 0.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.3 | 1.0 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 0.9 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 5.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.2 | 0.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 1.3 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 12.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 0.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.2 | 2.0 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.4 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.2 | 1.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 1.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.4 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 1.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 7.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.5 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 1.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.0 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.3 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 9.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 5.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 2.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 12.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.4 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.0 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.0 | 1.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |