Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
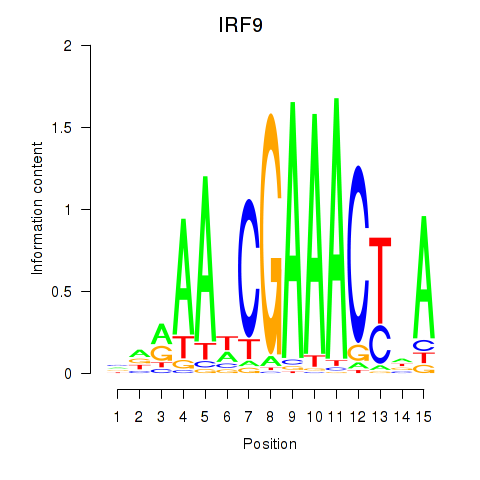
Results for IRF9
Z-value: 1.44
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF9
|
ENSG00000213928.4 | IRF9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF9 | hg19_v2_chr14_+_24630465_24630531 | 0.90 | 2.1e-06 | Click! |
Activity profile of IRF9 motif
Sorted Z-values of IRF9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_42798094 | 9.76 |
ENST00000398598.3 ENST00000455164.2 ENST00000424365.1 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr12_+_113344582 | 4.55 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_79086088 | 4.39 |
ENST00000370751.5 ENST00000342282.3 |
IFI44L |
interferon-induced protein 44-like |
| chr1_+_79115503 | 4.17 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr1_+_948803 | 4.09 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr12_+_113344755 | 4.01 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr6_+_6588902 | 3.64 |
ENST00000230568.4 |
LY86 |
lymphocyte antigen 86 |
| chr4_+_89378261 | 3.34 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr14_+_24630465 | 3.22 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr17_+_6659153 | 3.17 |
ENST00000441631.1 ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1 |
XIAP associated factor 1 |
| chr21_+_42733870 | 3.08 |
ENST00000330714.3 ENST00000436410.1 ENST00000435611.1 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
| chr1_+_158979792 | 3.06 |
ENST00000359709.3 ENST00000430894.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr6_-_31324943 | 2.90 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr1_+_158979680 | 2.90 |
ENST00000368131.4 ENST00000340979.6 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_-_615570 | 2.90 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr1_+_158979686 | 2.86 |
ENST00000368132.3 ENST00000295809.7 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr12_+_113344811 | 2.80 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_615942 | 2.52 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr2_-_163175133 | 2.49 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr4_-_169239921 | 2.47 |
ENST00000514995.1 ENST00000393743.3 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr9_-_21995249 | 2.43 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_-_21995300 | 2.34 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr14_+_94577074 | 2.33 |
ENST00000444961.1 ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27 |
interferon, alpha-inducible protein 27 |
| chr10_-_91174215 | 2.28 |
ENST00000371837.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr11_-_57335280 | 2.13 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr13_-_46961580 | 1.88 |
ENST00000378787.3 ENST00000378797.2 ENST00000429979.1 ENST00000378781.3 |
KIAA0226L |
KIAA0226-like |
| chr9_-_32526184 | 1.69 |
ENST00000545044.1 ENST00000379868.1 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr18_-_67624160 | 1.66 |
ENST00000581982.1 ENST00000280200.4 |
CD226 |
CD226 molecule |
| chr4_+_146402925 | 1.66 |
ENST00000302085.4 |
SMAD1 |
SMAD family member 1 |
| chr13_+_50070077 | 1.58 |
ENST00000378319.3 ENST00000426879.1 |
PHF11 |
PHD finger protein 11 |
| chr13_+_50070491 | 1.50 |
ENST00000496612.1 ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11 |
PHD finger protein 11 |
| chr11_+_5646213 | 1.40 |
ENST00000429814.2 |
TRIM34 |
tripartite motif containing 34 |
| chr4_-_76944621 | 1.33 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr12_+_113376157 | 1.32 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr10_-_95242044 | 1.13 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr10_-_95241951 | 1.12 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr1_-_150738261 | 1.11 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr10_+_91174314 | 1.03 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chr18_+_42260861 | 0.93 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr3_-_121379739 | 0.87 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr19_+_10196981 | 0.83 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr1_-_154580616 | 0.79 |
ENST00000368474.4 |
ADAR |
adenosine deaminase, RNA-specific |
| chr9_-_32526299 | 0.78 |
ENST00000379882.1 ENST00000379883.2 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr10_+_91092241 | 0.77 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_-_152146385 | 0.70 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr10_+_115439699 | 0.69 |
ENST00000369315.1 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr15_+_74287009 | 0.68 |
ENST00000395135.3 |
PML |
promyelocytic leukemia |
| chr1_+_241695424 | 0.65 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_+_115439282 | 0.64 |
ENST00000369321.2 ENST00000345633.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr4_+_37892682 | 0.62 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr10_+_115439630 | 0.61 |
ENST00000369318.3 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr1_+_241695670 | 0.59 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_-_4414880 | 0.58 |
ENST00000254436.7 ENST00000543625.1 |
TRIM21 |
tripartite motif containing 21 |
| chr6_-_33282163 | 0.57 |
ENST00000434618.2 ENST00000456592.2 |
TAPBP |
TAP binding protein (tapasin) |
| chr12_-_121477039 | 0.55 |
ENST00000257570.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chrX_+_37639264 | 0.54 |
ENST00000378588.4 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr9_+_33265011 | 0.48 |
ENST00000419016.2 |
CHMP5 |
charged multivesicular body protein 5 |
| chr22_+_18632666 | 0.47 |
ENST00000215794.7 |
USP18 |
ubiquitin specific peptidase 18 |
| chr3_-_146262352 | 0.46 |
ENST00000462666.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr17_-_40264692 | 0.46 |
ENST00000591220.1 ENST00000251642.3 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr16_-_15736881 | 0.45 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr3_-_146262637 | 0.45 |
ENST00000472349.1 ENST00000342435.4 |
PLSCR1 |
phospholipid scramblase 1 |
| chr17_+_7211656 | 0.44 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr9_+_33264861 | 0.44 |
ENST00000223500.8 |
CHMP5 |
charged multivesicular body protein 5 |
| chr6_+_106546808 | 0.41 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr22_+_40297079 | 0.34 |
ENST00000344138.4 ENST00000543252.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr10_+_91061712 | 0.32 |
ENST00000371826.3 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
| chr3_-_146262428 | 0.29 |
ENST00000486631.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chrX_-_63005405 | 0.27 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_-_37384175 | 0.25 |
ENST00000411537.2 ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2 |
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_-_146262488 | 0.25 |
ENST00000487389.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr3_-_146262365 | 0.25 |
ENST00000448787.2 |
PLSCR1 |
phospholipid scramblase 1 |
| chr12_+_6881678 | 0.24 |
ENST00000441671.2 ENST00000203629.2 |
LAG3 |
lymphocyte-activation gene 3 |
| chr5_-_142783694 | 0.22 |
ENST00000394466.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_-_67141090 | 0.19 |
ENST00000312438.7 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
| chrX_+_37639302 | 0.17 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr3_-_146262293 | 0.17 |
ENST00000448205.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr5_-_142783175 | 0.16 |
ENST00000231509.3 ENST00000394464.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_95297678 | 0.15 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr5_-_95297534 | 0.14 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr22_+_40297105 | 0.13 |
ENST00000540310.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr7_+_134832808 | 0.11 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr6_-_24936170 | 0.07 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr15_+_91643442 | 0.04 |
ENST00000394232.1 |
SV2B |
synaptic vesicle glycoprotein 2B |
| chr12_-_27167233 | 0.03 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.2 | 12.8 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 1.1 | 4.4 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.1 | 5.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.7 | 2.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.6 | 1.7 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.4 | 4.8 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.4 | 3.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 | 2.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 1.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 0.8 | GO:1900368 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.2 | 1.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 19.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 1.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 0.7 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.2 | 1.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 2.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.6 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.7 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 9.9 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.9 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 0.4 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.9 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 2.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 5.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 3.0 | GO:0009615 | response to virus(GO:0009615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.9 | 5.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 2.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.3 | 3.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 4.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 2.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 2.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 7.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 17.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.7 | GO:0050664 | superoxide-generating NADPH oxidase activity(GO:0016175) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 12.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 4.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 5.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.5 | 40.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 5.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 4.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.1 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 2.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 17.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 7.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.5 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |