Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for IRX3
Z-value: 1.21
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | IRX3 |
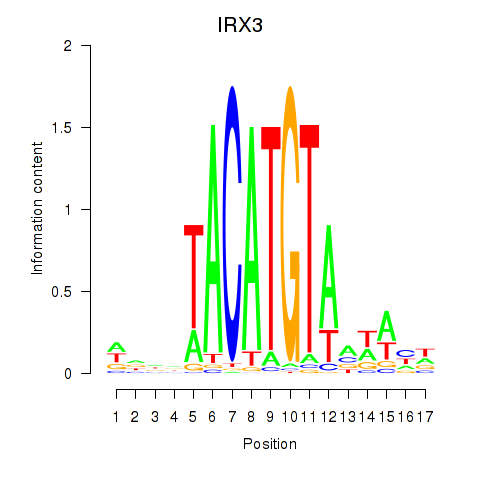
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_38172863 | 4.36 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr11_+_35201826 | 4.22 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr3_-_127541194 | 3.27 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr4_-_143481822 | 3.18 |
ENST00000510812.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_91574142 | 3.16 |
ENST00000547937.1 |
DCN |
decorin |
| chr11_+_35211429 | 2.99 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr15_+_49170083 | 2.84 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr12_-_91546926 | 2.69 |
ENST00000550758.1 |
DCN |
decorin |
| chr12_-_91573316 | 2.66 |
ENST00000393155.1 |
DCN |
decorin |
| chr12_-_91573132 | 2.66 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr4_+_169842707 | 2.56 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr11_-_35547572 | 2.49 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr12_-_91573249 | 2.23 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr5_+_35856951 | 2.19 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr3_+_8543393 | 2.19 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_189839046 | 2.17 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr1_+_158985457 | 2.16 |
ENST00000567661.1 ENST00000474473.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr11_+_114166536 | 2.15 |
ENST00000299964.3 |
NNMT |
nicotinamide N-methyltransferase |
| chr3_-_127441406 | 2.01 |
ENST00000487473.1 ENST00000484451.1 |
MGLL |
monoglyceride lipase |
| chr11_-_111782696 | 1.94 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr12_+_75874460 | 1.78 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr3_+_157154578 | 1.77 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr10_-_90712520 | 1.74 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr3_+_8543533 | 1.64 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr4_+_169418195 | 1.64 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_+_158979686 | 1.63 |
ENST00000368132.3 ENST00000295809.7 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr3_+_158787041 | 1.57 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr9_-_88715044 | 1.56 |
ENST00000388711.3 ENST00000466178.1 |
GOLM1 |
golgi membrane protein 1 |
| chr11_-_111782484 | 1.55 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr2_+_234621551 | 1.46 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr10_+_60272814 | 1.46 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr3_+_8543561 | 1.36 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr17_-_33760164 | 1.33 |
ENST00000445092.1 ENST00000394562.1 ENST00000447040.2 |
SLFN12 |
schlafen family member 12 |
| chr3_-_123512688 | 1.30 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr4_+_169418255 | 1.27 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr5_+_135385202 | 1.22 |
ENST00000514554.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr17_-_29648761 | 1.19 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr20_+_19867150 | 1.16 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr1_+_196788887 | 1.16 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr2_-_190044480 | 1.14 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr15_+_67420441 | 1.13 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr11_+_111783450 | 1.13 |
ENST00000537382.1 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr10_+_13628933 | 1.13 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr15_-_56757329 | 1.12 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr18_-_21891460 | 1.08 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr5_+_32788945 | 1.07 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr15_+_25200074 | 1.06 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr17_-_33760269 | 1.04 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr2_+_191792376 | 1.03 |
ENST00000409428.1 ENST00000409215.1 |
GLS |
glutaminase |
| chr8_-_101962777 | 1.03 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_144329531 | 1.02 |
ENST00000429150.1 ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr1_+_160175201 | 1.01 |
ENST00000368076.1 |
PEA15 |
phosphoprotein enriched in astrocytes 15 |
| chr1_+_160175117 | 1.01 |
ENST00000360472.4 |
PEA15 |
phosphoprotein enriched in astrocytes 15 |
| chr14_+_96722152 | 0.99 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr1_+_171060018 | 0.99 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr11_-_104905840 | 0.98 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr12_+_1738363 | 0.96 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr3_+_138066539 | 0.96 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr11_+_111782934 | 0.96 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr8_+_132952112 | 0.95 |
ENST00000520362.1 ENST00000519656.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr15_+_25200108 | 0.94 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr9_-_21975088 | 0.91 |
ENST00000304494.5 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr18_-_53303123 | 0.87 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr10_-_103815874 | 0.87 |
ENST00000370033.4 ENST00000311122.5 |
C10orf76 |
chromosome 10 open reading frame 76 |
| chr2_+_233527443 | 0.85 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr11_+_60223225 | 0.84 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr12_+_56114151 | 0.84 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr13_+_73629107 | 0.83 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr7_+_40174565 | 0.81 |
ENST00000309930.5 ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10 |
succinylCoA:glutarate-CoA transferase |
| chr1_-_154909329 | 0.81 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr12_+_56114189 | 0.81 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr9_+_4679555 | 0.80 |
ENST00000381858.1 ENST00000381854.3 |
CDC37L1 |
cell division cycle 37-like 1 |
| chr2_-_16804320 | 0.80 |
ENST00000355549.2 |
FAM49A |
family with sequence similarity 49, member A |
| chr17_-_29641104 | 0.80 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr11_-_14358620 | 0.79 |
ENST00000531421.1 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
| chr1_+_87458692 | 0.77 |
ENST00000370548.2 ENST00000356813.4 |
RP5-1052I5.2 HS2ST1 |
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr16_+_56969284 | 0.76 |
ENST00000568358.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr17_+_18601299 | 0.75 |
ENST00000572555.1 ENST00000395902.3 ENST00000449552.2 |
TRIM16L |
tripartite motif containing 16-like |
| chr11_-_104972158 | 0.74 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr19_-_22379753 | 0.74 |
ENST00000397121.2 |
ZNF676 |
zinc finger protein 676 |
| chr8_-_108510224 | 0.71 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr2_-_158300556 | 0.71 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr12_-_52867569 | 0.71 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr7_-_41742697 | 0.71 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr10_+_13628921 | 0.70 |
ENST00000378572.3 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr6_-_32920794 | 0.66 |
ENST00000395305.3 ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA XXbac-BPG181M17.5 |
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr1_-_62190793 | 0.66 |
ENST00000371177.2 ENST00000606498.1 |
TM2D1 |
TM2 domain containing 1 |
| chr11_+_86013253 | 0.65 |
ENST00000533986.1 ENST00000278483.3 |
C11orf73 |
chromosome 11 open reading frame 73 |
| chr19_-_43708378 | 0.65 |
ENST00000599746.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr12_+_5541267 | 0.65 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr11_-_61647935 | 0.65 |
ENST00000531956.1 |
FADS3 |
fatty acid desaturase 3 |
| chr2_-_231090344 | 0.65 |
ENST00000540870.1 ENST00000416610.1 |
SP110 |
SP110 nuclear body protein |
| chr17_-_29641084 | 0.64 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr16_+_50300427 | 0.64 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr12_-_10007448 | 0.64 |
ENST00000538152.1 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr2_-_86094764 | 0.63 |
ENST00000393808.3 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr1_+_144173162 | 0.63 |
ENST00000356801.6 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr4_+_41361616 | 0.63 |
ENST00000513024.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr7_+_44240520 | 0.62 |
ENST00000496112.1 ENST00000223369.2 |
YKT6 |
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr1_+_160709055 | 0.60 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr1_-_9129735 | 0.59 |
ENST00000377424.4 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_-_23504176 | 0.58 |
ENST00000302291.4 |
LUZP1 |
leucine zipper protein 1 |
| chr2_+_234637754 | 0.58 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_-_170924888 | 0.58 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr4_-_70826725 | 0.57 |
ENST00000353151.3 |
CSN2 |
casein beta |
| chr4_+_41992489 | 0.57 |
ENST00000264451.7 |
SLC30A9 |
solute carrier family 30 (zinc transporter), member 9 |
| chrX_+_99899180 | 0.57 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr8_+_125463048 | 0.56 |
ENST00000328599.3 |
TRMT12 |
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr7_+_120591170 | 0.55 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr1_+_196912902 | 0.55 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr1_+_145524891 | 0.55 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr13_-_36429763 | 0.54 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr12_+_16109519 | 0.54 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr17_-_63822563 | 0.53 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr14_-_106926724 | 0.53 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr2_-_113594279 | 0.53 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr5_+_140810132 | 0.53 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr1_+_160709029 | 0.52 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chr14_-_106692191 | 0.51 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr15_+_25101698 | 0.51 |
ENST00000400097.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_48936272 | 0.50 |
ENST00000544097.1 ENST00000430379.1 ENST00000319017.4 |
SLC25A20 |
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr2_-_130886795 | 0.50 |
ENST00000409914.2 |
POTEF |
POTE ankyrin domain family, member F |
| chr14_-_106845789 | 0.49 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr1_-_230513367 | 0.48 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr14_+_96722539 | 0.48 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr10_+_91092241 | 0.47 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_+_160590469 | 0.47 |
ENST00000409591.1 |
MARCH7 |
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr11_-_87908600 | 0.47 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr4_+_75480629 | 0.47 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr7_+_116502527 | 0.46 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr12_+_21590549 | 0.46 |
ENST00000545178.1 ENST00000240651.9 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr6_-_32634425 | 0.45 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr6_-_111804393 | 0.45 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr14_-_67878917 | 0.45 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr15_-_81616446 | 0.44 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr1_+_155829286 | 0.44 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr9_-_135230336 | 0.44 |
ENST00000224140.5 ENST00000372169.2 ENST00000393220.1 |
SETX |
senataxin |
| chr1_-_148025848 | 0.44 |
ENST00000310701.10 ENST00000369219.1 ENST00000444640.1 ENST00000431121.1 ENST00000436356.1 ENST00000448574.1 ENST00000458135.1 ENST00000392972.3 ENST00000426874.1 |
NBPF14 |
neuroblastoma breakpoint family, member 14 |
| chr16_-_33647696 | 0.44 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr7_+_144052381 | 0.44 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_106963409 | 0.44 |
ENST00000390621.2 |
IGHV1-45 |
immunoglobulin heavy variable 1-45 |
| chr16_+_33605231 | 0.43 |
ENST00000570121.2 |
IGHV3OR16-12 |
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr5_-_93447333 | 0.43 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr14_-_106471723 | 0.43 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr9_-_95298314 | 0.42 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr3_+_122044084 | 0.42 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr1_+_144151520 | 0.41 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr6_+_29426230 | 0.41 |
ENST00000442615.1 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr6_+_127587755 | 0.41 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr1_-_201438282 | 0.41 |
ENST00000367311.3 ENST00000367309.1 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
| chr1_-_197036364 | 0.41 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr1_+_53480598 | 0.40 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr6_-_154677900 | 0.39 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr18_+_21529811 | 0.39 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr9_-_95244781 | 0.39 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr8_+_22429205 | 0.39 |
ENST00000520207.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr7_+_80267973 | 0.39 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_-_21757774 | 0.38 |
ENST00000261195.2 |
GYS2 |
glycogen synthase 2 (liver) |
| chr16_+_81070792 | 0.38 |
ENST00000564241.1 ENST00000565237.1 |
ATMIN |
ATM interactor |
| chr4_+_75310851 | 0.38 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr6_-_117747015 | 0.37 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr19_-_20748541 | 0.37 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr1_-_46598371 | 0.37 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr4_+_88754069 | 0.37 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr1_+_87012753 | 0.37 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr3_+_142838091 | 0.37 |
ENST00000309575.3 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr7_+_55086703 | 0.37 |
ENST00000455089.1 ENST00000342916.3 ENST00000344576.2 ENST00000420316.2 |
EGFR |
epidermal growth factor receptor |
| chr18_-_28622774 | 0.37 |
ENST00000434452.1 |
DSC3 |
desmocollin 3 |
| chr14_-_51562745 | 0.36 |
ENST00000298355.3 |
TRIM9 |
tripartite motif containing 9 |
| chr9_-_13279589 | 0.36 |
ENST00000319217.7 |
MPDZ |
multiple PDZ domain protein |
| chr13_+_21714653 | 0.35 |
ENST00000382533.4 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr7_+_77469439 | 0.35 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr2_+_234590556 | 0.35 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr16_-_30032610 | 0.35 |
ENST00000574405.1 |
DOC2A |
double C2-like domains, alpha |
| chr10_-_112678976 | 0.34 |
ENST00000448814.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr3_-_27498235 | 0.34 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_-_115124257 | 0.34 |
ENST00000369541.3 |
BCAS2 |
breast carcinoma amplified sequence 2 |
| chrX_-_15402498 | 0.34 |
ENST00000297904.3 |
FIGF |
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr1_+_202317815 | 0.33 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_115420688 | 0.33 |
ENST00000274458.4 |
COMMD10 |
COMM domain containing 10 |
| chr3_+_186383741 | 0.32 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr21_-_27462351 | 0.32 |
ENST00000448850.1 |
APP |
amyloid beta (A4) precursor protein |
| chr1_+_201924619 | 0.32 |
ENST00000367287.4 |
TIMM17A |
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr1_+_77333117 | 0.32 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr19_-_45004556 | 0.31 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr4_-_69434245 | 0.31 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr11_+_114310237 | 0.31 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr2_-_86790593 | 0.31 |
ENST00000263856.4 ENST00000409225.2 |
CHMP3 |
charged multivesicular body protein 3 |
| chr3_-_50378235 | 0.30 |
ENST00000357043.2 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr3_-_185641681 | 0.30 |
ENST00000259043.7 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
| chr1_-_114414316 | 0.30 |
ENST00000528414.1 ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr11_+_114310102 | 0.30 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr12_-_102224704 | 0.30 |
ENST00000299314.7 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr3_-_57583185 | 0.29 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_+_132316081 | 0.29 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr6_-_32784687 | 0.29 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr11_-_26743546 | 0.29 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr15_-_63448973 | 0.29 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr5_-_9630463 | 0.29 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr1_+_196857144 | 0.28 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.0 | 13.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 1.8 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.4 | 1.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.7 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 0.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 3.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 1.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 5.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 3.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 4.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0046696 | CBM complex(GO:0032449) lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.0 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 5.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 6.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 3.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 5.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.1 | 13.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.9 | 7.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.8 | 5.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 | 1.8 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.4 | 3.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 0.8 | GO:0032943 | mononuclear cell proliferation(GO:0032943) lymphocyte proliferation(GO:0046651) leukocyte proliferation(GO:0070661) |
| 0.4 | 2.0 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.4 | 0.4 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.4 | 0.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 | 1.6 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 1.7 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.3 | 1.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 5.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.6 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 1.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 5.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 2.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 | 3.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.6 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.2 | 0.7 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.2 | 0.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.2 | 0.5 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.2 | 1.8 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.4 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 2.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 1.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.5 | GO:0048871 | multicellular organismal homeostasis(GO:0048871) |
| 0.1 | 0.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.2 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.1 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.1 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.6 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 4.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.4 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.2 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 1.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.4 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) coagulation(GO:0050817) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.4 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.2 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.1 | 3.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 3.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0018212 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 2.8 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 1.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.3 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:1903593 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.4 | GO:0016032 | viral process(GO:0016032) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0009914 | hormone transport(GO:0009914) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.3 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.0 | 0.1 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.0 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0042127 | regulation of cell proliferation(GO:0042127) |
| 0.0 | 0.7 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0030031 | cell projection assembly(GO:0030031) |
| 0.0 | 0.1 | GO:2000255 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.0 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 1.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.0 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.1 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.6 | 3.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 1.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.5 | 2.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.5 | 1.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 5.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 24.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 5.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.5 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 0.5 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.2 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 3.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 3.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.8 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 3.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 4.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 2.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |