Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ISL2
Z-value: 0.66
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL2 |
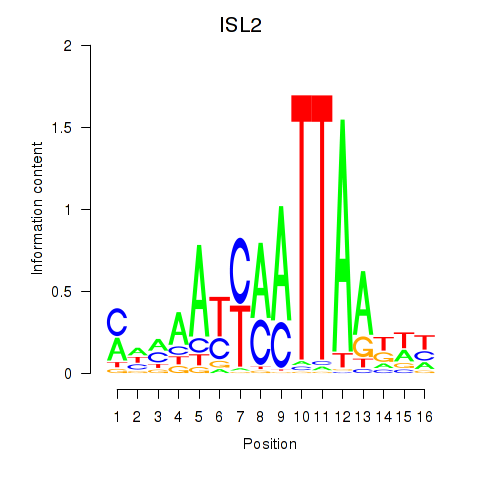
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95241951 | 2.98 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr10_-_95242044 | 2.95 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr4_+_169418195 | 1.35 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr14_-_67878917 | 1.28 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr11_-_62521614 | 1.25 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr11_+_12132117 | 1.10 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_+_234668894 | 1.05 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_99127225 | 0.97 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr4_-_90756769 | 0.93 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_+_169418255 | 0.87 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr21_+_39644395 | 0.86 |
ENST00000398934.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_5255861 | 0.86 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr21_+_39644305 | 0.83 |
ENST00000398930.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_-_139726181 | 0.73 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr11_-_107729887 | 0.72 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr17_-_7307358 | 0.60 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_234580525 | 0.59 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_+_136649311 | 0.56 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr2_+_234580499 | 0.56 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr3_-_127455200 | 0.52 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chr18_-_53303123 | 0.52 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr15_-_72563585 | 0.50 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr1_+_77333117 | 0.50 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr15_+_64680003 | 0.48 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chrX_+_65382433 | 0.47 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chrX_+_65384182 | 0.46 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr2_-_10587897 | 0.45 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr2_-_87248975 | 0.35 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr4_+_169013666 | 0.35 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chrX_-_48931648 | 0.34 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr8_+_70404996 | 0.34 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr15_+_79166065 | 0.31 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr3_-_141747950 | 0.31 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_327537 | 0.30 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr2_-_136633940 | 0.29 |
ENST00000264156.2 |
MCM6 |
minichromosome maintenance complex component 6 |
| chr2_+_231921574 | 0.28 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr2_-_145277569 | 0.27 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_54689532 | 0.27 |
ENST00000540264.2 ENST00000312156.4 |
NFE2 |
nuclear factor, erythroid 2 |
| chrX_-_100872911 | 0.26 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr2_+_88047606 | 0.26 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr4_-_120243545 | 0.24 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr8_-_121457608 | 0.23 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr1_-_93257951 | 0.23 |
ENST00000543509.1 ENST00000370331.1 ENST00000540033.1 |
EVI5 |
ecotropic viral integration site 5 |
| chr10_-_29923893 | 0.23 |
ENST00000355867.4 |
SVIL |
supervillin |
| chrX_+_55478538 | 0.22 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr2_-_217560248 | 0.22 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr18_-_64271363 | 0.21 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr12_+_26164645 | 0.20 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr19_+_15852203 | 0.19 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr14_+_38677123 | 0.19 |
ENST00000267377.2 |
SSTR1 |
somatostatin receptor 1 |
| chr7_-_115670804 | 0.18 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr3_-_164796269 | 0.17 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chrX_+_102840408 | 0.17 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr7_-_115670792 | 0.15 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr5_+_32788945 | 0.15 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr1_-_228613026 | 0.14 |
ENST00000366696.1 |
HIST3H3 |
histone cluster 3, H3 |
| chr8_-_30706608 | 0.14 |
ENST00000256246.2 |
TEX15 |
testis expressed 15 |
| chr1_-_202897724 | 0.13 |
ENST00000435533.3 ENST00000367258.1 |
KLHL12 |
kelch-like family member 12 |
| chr14_-_23058063 | 0.11 |
ENST00000538631.1 ENST00000543337.1 ENST00000250498.4 |
DAD1 |
defender against cell death 1 |
| chr1_-_95391315 | 0.11 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr3_-_183145873 | 0.09 |
ENST00000447025.2 ENST00000414362.2 ENST00000328913.3 |
MCF2L2 |
MCF.2 cell line derived transforming sequence-like 2 |
| chr17_-_10325261 | 0.09 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr2_-_217559517 | 0.09 |
ENST00000449583.1 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr12_+_75784850 | 0.09 |
ENST00000550916.1 ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2 |
GLI pathogenesis-related 1 like 2 |
| chr14_-_106781017 | 0.09 |
ENST00000390612.2 |
IGHV4-28 |
immunoglobulin heavy variable 4-28 |
| chr6_+_151561085 | 0.08 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr14_-_51027838 | 0.08 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chrX_-_84634737 | 0.08 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr3_+_172468505 | 0.08 |
ENST00000427830.1 ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr3_+_172468472 | 0.08 |
ENST00000232458.5 ENST00000392692.3 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr3_+_139063372 | 0.07 |
ENST00000478464.1 |
MRPS22 |
mitochondrial ribosomal protein S22 |
| chr6_+_26087509 | 0.07 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr19_+_4402659 | 0.07 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr1_+_63063152 | 0.07 |
ENST00000371129.3 |
ANGPTL3 |
angiopoietin-like 3 |
| chr3_+_138327417 | 0.07 |
ENST00000338446.4 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr1_-_174992544 | 0.07 |
ENST00000476371.1 |
MRPS14 |
mitochondrial ribosomal protein S14 |
| chr1_+_149871135 | 0.06 |
ENST00000369152.5 |
BOLA1 |
bolA family member 1 |
| chr11_+_113779704 | 0.06 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr4_+_128702969 | 0.06 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr3_+_138327542 | 0.06 |
ENST00000360570.3 ENST00000393035.2 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr5_-_130500922 | 0.06 |
ENST00000513012.1 ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1 |
histidine triad nucleotide binding protein 1 |
| chr12_-_10282836 | 0.05 |
ENST00000304084.8 ENST00000353231.5 ENST00000525605.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr4_+_113568207 | 0.05 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr9_-_95166841 | 0.05 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr2_-_209010874 | 0.04 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chrX_-_77225135 | 0.04 |
ENST00000458128.1 |
PGAM4 |
phosphoglycerate mutase family member 4 |
| chr15_-_42500351 | 0.04 |
ENST00000348544.4 ENST00000318006.5 |
VPS39 |
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr4_+_88532028 | 0.04 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr9_+_111624577 | 0.03 |
ENST00000333999.3 |
ACTL7A |
actin-like 7A |
| chr18_-_3845293 | 0.03 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_169887827 | 0.03 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr12_+_4829507 | 0.03 |
ENST00000252318.2 |
GALNT8 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8) |
| chr12_-_118628350 | 0.03 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr11_+_86085778 | 0.03 |
ENST00000354755.1 ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81 |
coiled-coil domain containing 81 |
| chr9_+_12693336 | 0.03 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr14_-_90798418 | 0.02 |
ENST00000354366.3 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
| chr19_-_51920952 | 0.02 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr17_-_10372875 | 0.01 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr22_-_29107919 | 0.01 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr4_+_71108300 | 0.01 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr11_+_73498898 | 0.01 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chrX_+_11311533 | 0.01 |
ENST00000380714.3 ENST00000380712.3 ENST00000348912.4 |
AMELX |
amelogenin, X-linked |
| chrX_+_96138907 | 0.01 |
ENST00000373040.3 |
RPA4 |
replication protein A4, 30kDa |
| chr1_+_149871171 | 0.00 |
ENST00000369150.1 |
BOLA1 |
bolA family member 1 |
| chr18_-_3845321 | 0.00 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_-_190446759 | 0.00 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 5.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 6.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 1.6 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.2 | 0.6 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.2 | 0.9 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.6 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 2.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 1.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |