Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
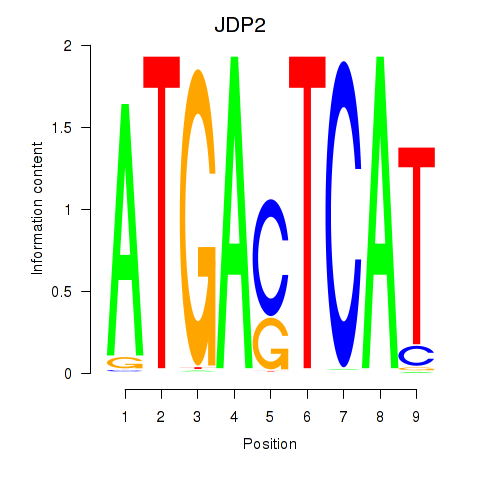
Results for JDP2
Z-value: 0.95
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | JDP2 |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_151965048 | 4.19 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr2_-_175712270 | 4.06 |
ENST00000295497.7 ENST00000444394.1 |
CHN1 |
chimerin 1 |
| chr11_-_102668879 | 3.26 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr7_+_134528635 | 3.09 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr5_+_135394840 | 2.92 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr8_-_27472198 | 2.42 |
ENST00000519472.1 ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU |
clusterin |
| chr3_-_32022733 | 2.19 |
ENST00000438237.2 ENST00000396556.2 |
OSBPL10 |
oxysterol binding protein-like 10 |
| chr11_-_2950642 | 2.07 |
ENST00000314222.4 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
| chrY_+_15016725 | 1.82 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr12_-_54813229 | 1.73 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr3_-_49395705 | 1.68 |
ENST00000419349.1 |
GPX1 |
glutathione peroxidase 1 |
| chr16_-_69760409 | 1.55 |
ENST00000561500.1 ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1 |
NAD(P)H dehydrogenase, quinone 1 |
| chr5_+_35856951 | 1.48 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr11_+_101983176 | 1.46 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr10_+_17270214 | 1.40 |
ENST00000544301.1 |
VIM |
vimentin |
| chr7_-_80551671 | 1.38 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_+_10365404 | 1.26 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr16_+_15596123 | 1.23 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr3_-_149293990 | 1.14 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr3_-_114343768 | 1.14 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr9_-_35112376 | 1.13 |
ENST00000488109.2 |
FAM214B |
family with sequence similarity 214, member B |
| chr7_+_55177416 | 1.10 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr8_+_70404996 | 1.05 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr7_+_48128194 | 1.00 |
ENST00000416681.1 ENST00000331803.4 ENST00000432131.1 |
UPP1 |
uridine phosphorylase 1 |
| chr7_+_48128316 | 0.99 |
ENST00000341253.4 |
UPP1 |
uridine phosphorylase 1 |
| chr9_-_21335356 | 0.96 |
ENST00000359039.4 |
KLHL9 |
kelch-like family member 9 |
| chr10_+_121410882 | 0.90 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr17_-_33760269 | 0.87 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr1_+_223889285 | 0.86 |
ENST00000433674.2 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr9_-_21335240 | 0.81 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr3_+_69134124 | 0.81 |
ENST00000478935.1 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr8_-_82395461 | 0.76 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr15_+_22892663 | 0.74 |
ENST00000313077.7 ENST00000561274.1 ENST00000560848.1 |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
| chr3_+_69134080 | 0.72 |
ENST00000273258.3 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr16_+_57662419 | 0.72 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57662138 | 0.70 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr10_+_1102721 | 0.69 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr20_+_4666882 | 0.68 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr6_-_56819385 | 0.66 |
ENST00000370754.5 ENST00000449297.2 |
DST |
dystonin |
| chr4_+_74606223 | 0.65 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr13_+_53602894 | 0.63 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr15_-_60690163 | 0.62 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr3_-_18466787 | 0.59 |
ENST00000338745.6 ENST00000450898.1 |
SATB1 |
SATB homeobox 1 |
| chrX_-_48901012 | 0.58 |
ENST00000315869.7 |
TFE3 |
transcription factor binding to IGHM enhancer 3 |
| chr8_+_15397732 | 0.58 |
ENST00000382020.4 ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3 |
tumor suppressor candidate 3 |
| chr2_-_145275228 | 0.57 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr3_-_122512619 | 0.56 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chr11_-_62323702 | 0.56 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr7_+_23286182 | 0.56 |
ENST00000258733.4 ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB |
glycoprotein (transmembrane) nmb |
| chr11_-_102401469 | 0.54 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_+_26606608 | 0.53 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chrX_-_15288154 | 0.50 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chr7_+_872107 | 0.49 |
ENST00000405266.1 ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
| chr22_+_39916558 | 0.47 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr3_-_18466026 | 0.47 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr17_+_35851570 | 0.46 |
ENST00000394386.1 |
DUSP14 |
dual specificity phosphatase 14 |
| chr1_+_156084461 | 0.44 |
ENST00000347559.2 ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA |
lamin A/C |
| chr16_+_89988259 | 0.41 |
ENST00000554444.1 ENST00000556565.1 |
TUBB3 |
Tubulin beta-3 chain |
| chr2_+_152214098 | 0.39 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr1_-_160231451 | 0.38 |
ENST00000495887.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr19_-_46285646 | 0.37 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr19_+_3366547 | 0.37 |
ENST00000341919.3 ENST00000590282.1 ENST00000443272.2 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr12_-_56101647 | 0.37 |
ENST00000347027.6 ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7 |
integrin, alpha 7 |
| chr17_+_79650962 | 0.37 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr1_+_156096336 | 0.36 |
ENST00000504687.1 ENST00000473598.2 |
LMNA |
lamin A/C |
| chr1_-_95007193 | 0.36 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr4_+_169013666 | 0.32 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr21_-_36421535 | 0.32 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr6_-_84140757 | 0.32 |
ENST00000541327.1 ENST00000369705.3 ENST00000543031.1 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr5_+_179247759 | 0.31 |
ENST00000389805.4 ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1 |
sequestosome 1 |
| chr9_-_23826298 | 0.29 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr19_-_4455290 | 0.29 |
ENST00000394765.3 ENST00000592515.1 |
UBXN6 |
UBX domain protein 6 |
| chr2_+_46926048 | 0.28 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr21_-_36421626 | 0.28 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr11_-_66103867 | 0.28 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr8_-_141774467 | 0.26 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr17_+_75315534 | 0.26 |
ENST00000590294.1 ENST00000329047.8 |
SEPT9 |
septin 9 |
| chr13_+_32838801 | 0.26 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chrX_+_153770421 | 0.25 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr12_+_57914742 | 0.25 |
ENST00000551351.1 |
MBD6 |
methyl-CpG binding domain protein 6 |
| chr2_+_28615669 | 0.25 |
ENST00000379619.1 ENST00000264716.4 |
FOSL2 |
FOS-like antigen 2 |
| chr11_-_75201791 | 0.25 |
ENST00000529721.1 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr4_+_120056939 | 0.24 |
ENST00000307128.5 |
MYOZ2 |
myozenin 2 |
| chr12_-_57914275 | 0.22 |
ENST00000547303.1 ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3 |
DNA-damage-inducible transcript 3 |
| chr19_-_45927622 | 0.22 |
ENST00000300853.3 ENST00000589165.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr5_-_138534071 | 0.22 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr12_-_48152611 | 0.19 |
ENST00000389212.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_-_102826434 | 0.19 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr19_+_13842559 | 0.17 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr4_-_987217 | 0.17 |
ENST00000361661.2 ENST00000398516.2 |
SLC26A1 |
solute carrier family 26 (anion exchanger), member 1 |
| chr21_-_36421401 | 0.16 |
ENST00000486278.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr19_-_5903714 | 0.16 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr1_-_244006528 | 0.15 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_+_72143988 | 0.15 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr17_-_73775839 | 0.15 |
ENST00000592643.1 ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B |
H3 histone, family 3B (H3.3B) |
| chr5_-_149492904 | 0.14 |
ENST00000286301.3 ENST00000511344.1 |
CSF1R |
colony stimulating factor 1 receptor |
| chr13_-_36429763 | 0.13 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr11_-_102714534 | 0.13 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr5_-_148929848 | 0.12 |
ENST00000504676.1 ENST00000515435.1 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chr6_+_63921351 | 0.12 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chrX_+_1734051 | 0.12 |
ENST00000381229.4 ENST00000381233.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr12_+_6644443 | 0.12 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr22_-_39715600 | 0.12 |
ENST00000427905.1 ENST00000402527.1 ENST00000216146.4 |
RPL3 |
ribosomal protein L3 |
| chr9_+_118950325 | 0.11 |
ENST00000534838.1 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr3_+_48507621 | 0.11 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chrX_+_1733876 | 0.11 |
ENST00000381241.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr1_+_213224572 | 0.11 |
ENST00000543470.1 ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr12_+_107168342 | 0.11 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr3_+_48507210 | 0.10 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr12_-_48152853 | 0.10 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_-_66103932 | 0.10 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr4_+_175839551 | 0.10 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr16_+_28996364 | 0.09 |
ENST00000564277.1 |
LAT |
linker for activation of T cells |
| chr3_-_194090460 | 0.08 |
ENST00000428839.1 ENST00000347624.3 |
LRRC15 |
leucine rich repeat containing 15 |
| chr12_+_107168418 | 0.08 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr9_+_140135665 | 0.08 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr12_+_52695617 | 0.07 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr16_+_3692931 | 0.07 |
ENST00000407479.1 |
DNASE1 |
deoxyribonuclease I |
| chr20_-_17539456 | 0.06 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr17_-_53800217 | 0.06 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr11_-_82708519 | 0.06 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr20_+_30946106 | 0.05 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr1_-_45476944 | 0.05 |
ENST00000372172.4 |
HECTD3 |
HECT domain containing E3 ubiquitin protein ligase 3 |
| chr9_-_74979420 | 0.05 |
ENST00000343431.2 ENST00000376956.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr14_+_73525144 | 0.05 |
ENST00000261973.7 ENST00000540173.1 |
RBM25 |
RNA binding motif protein 25 |
| chr17_-_47785504 | 0.05 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr4_-_122085469 | 0.05 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr4_+_3388057 | 0.05 |
ENST00000538395.1 |
RGS12 |
regulator of G-protein signaling 12 |
| chr1_+_17634689 | 0.04 |
ENST00000375453.1 ENST00000375448.4 |
PADI4 |
peptidyl arginine deiminase, type IV |
| chr1_-_111148241 | 0.04 |
ENST00000440270.1 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr19_+_50706866 | 0.04 |
ENST00000440075.2 ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14 |
myosin, heavy chain 14, non-muscle |
| chr10_+_88428206 | 0.04 |
ENST00000429277.2 ENST00000458213.2 ENST00000352360.5 |
LDB3 |
LIM domain binding 3 |
| chr17_+_66245341 | 0.04 |
ENST00000577985.1 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr17_+_48712206 | 0.04 |
ENST00000427699.1 ENST00000285238.8 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr6_-_161695042 | 0.04 |
ENST00000366908.5 ENST00000366911.5 ENST00000366905.3 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_+_27719148 | 0.04 |
ENST00000374024.3 |
GPR3 |
G protein-coupled receptor 3 |
| chr1_-_93257951 | 0.03 |
ENST00000543509.1 ENST00000370331.1 ENST00000540033.1 |
EVI5 |
ecotropic viral integration site 5 |
| chr18_+_21693306 | 0.03 |
ENST00000540918.2 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr11_-_2924720 | 0.03 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_-_10324716 | 0.02 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr16_+_31366455 | 0.02 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_+_154377669 | 0.01 |
ENST00000368485.3 ENST00000344086.4 |
IL6R |
interleukin 6 receptor |
| chr16_+_28996416 | 0.01 |
ENST00000395456.2 ENST00000454369.2 |
LAT |
linker for activation of T cells |
| chr17_+_74381343 | 0.01 |
ENST00000392496.3 |
SPHK1 |
sphingosine kinase 1 |
| chr11_-_82708435 | 0.00 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr4_+_175839506 | 0.00 |
ENST00000505141.1 ENST00000359240.3 ENST00000445694.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr5_-_138533973 | 0.00 |
ENST00000507002.1 ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr7_-_55606346 | 0.00 |
ENST00000545390.1 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.5 | 1.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 2.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 3.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 3.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.4 | 2.4 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 1.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 1.7 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 1.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 4.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 1.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 2.6 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 0.8 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.4 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 1.7 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 2.1 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 3.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 1.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 1.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 4.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.6 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 1.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 1.5 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.4 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.2 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 2.5 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 1.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.3 | 1.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 2.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 3.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.5 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0031307 | nuclear outer membrane(GO:0005640) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 3.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |