Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
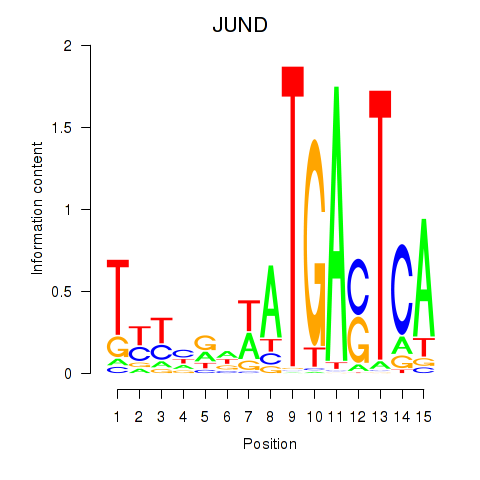
Results for JUND
Z-value: 1.32
Transcription factors associated with JUND
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUND
|
ENSG00000130522.4 | JUND |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUND | hg19_v2_chr19_-_18392422_18392440 | 0.56 | 2.3e-02 | Click! |
Activity profile of JUND motif
Sorted Z-values of JUND motif
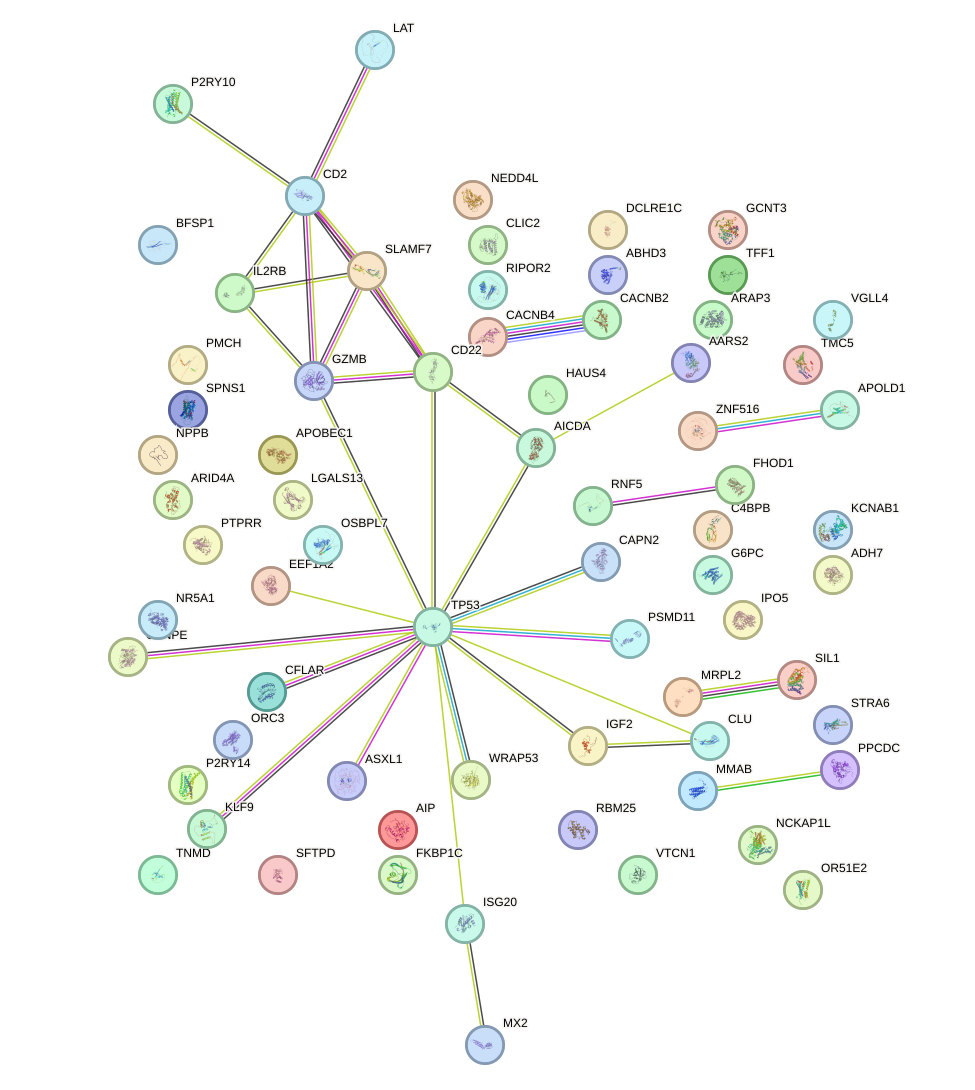
Network of associatons between targets according to the STRING database.
First level regulatory network of JUND
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_8765446 | 4.25 |
ENST00000537228.1 ENST00000229335.6 |
AICDA |
activation-induced cytidine deaminase |
| chr4_-_84035868 | 3.91 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr4_-_84035905 | 3.82 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr16_+_23847339 | 3.72 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr22_-_37545972 | 3.72 |
ENST00000216223.5 |
IL2RB |
interleukin 2 receptor, beta |
| chr1_+_160709029 | 2.49 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chrX_-_154563889 | 2.35 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr1_+_160709055 | 2.30 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr15_+_89181974 | 2.17 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chrX_+_78200829 | 2.12 |
ENST00000544091.1 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr12_+_7023491 | 1.96 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr22_+_23101182 | 1.95 |
ENST00000390312.2 |
IGLV2-14 |
immunoglobulin lambda variable 2-14 |
| chr15_+_89182178 | 1.95 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 1.94 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr11_+_67250490 | 1.91 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr18_-_74207146 | 1.87 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr21_+_42733870 | 1.74 |
ENST00000330714.3 ENST00000436410.1 ENST00000435611.1 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
| chr1_+_223889310 | 1.65 |
ENST00000434648.1 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr11_-_2162162 | 1.62 |
ENST00000381389.1 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr3_-_105588231 | 1.58 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_54891495 | 1.28 |
ENST00000293373.6 |
NCKAP1L |
NCK-associated protein 1-like |
| chr8_-_17752912 | 1.28 |
ENST00000398054.1 ENST00000381840.2 |
FGL1 |
fibrinogen-like 1 |
| chr6_+_32146131 | 1.27 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr14_-_23426270 | 1.17 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426337 | 1.16 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr8_-_17752996 | 1.16 |
ENST00000381841.2 ENST00000427924.1 |
FGL1 |
fibrinogen-like 1 |
| chr14_-_23426322 | 1.15 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr11_-_2162468 | 1.13 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr3_-_98241760 | 1.11 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chrX_-_51797351 | 1.10 |
ENST00000375644.3 |
RP11-114H20.1 |
RP11-114H20.1 |
| chr20_+_30946106 | 1.09 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chrY_+_15016725 | 1.07 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrX_+_51942963 | 0.96 |
ENST00000375625.3 |
RP11-363G10.2 |
RP11-363G10.2 |
| chr15_+_75335604 | 0.93 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr15_+_59903975 | 0.91 |
ENST00000560585.1 ENST00000396065.1 |
GCNT3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_+_207262881 | 0.89 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr19_+_35820064 | 0.87 |
ENST00000341773.6 ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22 |
CD22 molecule |
| chr17_+_41052808 | 0.87 |
ENST00000592383.1 ENST00000253801.2 ENST00000585489.1 |
G6PC |
glucose-6-phosphatase, catalytic subunit |
| chr1_-_11918988 | 0.85 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr4_-_100356291 | 0.81 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_+_76311791 | 0.81 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr12_-_102591604 | 0.78 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr21_-_43786634 | 0.77 |
ENST00000291527.2 |
TFF1 |
trefoil factor 1 |
| chr7_+_33765593 | 0.74 |
ENST00000311067.3 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
| chr16_+_31366536 | 0.72 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr11_-_82708435 | 0.72 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr4_-_123542224 | 0.69 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr8_-_27472198 | 0.68 |
ENST00000519472.1 ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU |
clusterin |
| chr20_-_17539456 | 0.66 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr12_+_12938541 | 0.65 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr6_-_44281043 | 0.64 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr1_-_54637997 | 0.61 |
ENST00000536061.1 |
AL357673.1 |
CDNA: FLJ21031 fis, clone CAE07336; HCG1780521; Uncharacterized protein |
| chr18_-_19283649 | 0.58 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr17_+_7590734 | 0.56 |
ENST00000457584.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr13_+_98605902 | 0.55 |
ENST00000460070.1 ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5 |
importin 5 |
| chr16_+_28996114 | 0.54 |
ENST00000395461.3 |
LAT |
linker for activation of T cells |
| chr2_+_169926047 | 0.54 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr17_-_45899126 | 0.53 |
ENST00000007414.3 ENST00000392507.3 |
OSBPL7 |
oxysterol binding protein-like 7 |
| chr4_-_100356551 | 0.53 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_43027105 | 0.53 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr6_+_44194762 | 0.50 |
ENST00000371708.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr6_-_24936170 | 0.49 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr12_-_71182695 | 0.48 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr5_-_141061759 | 0.47 |
ENST00000508305.1 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr15_-_74504560 | 0.46 |
ENST00000449139.2 |
STRA6 |
stimulated by retinoic acid 6 |
| chr4_-_104119528 | 0.46 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chr11_-_5255696 | 0.44 |
ENST00000292901.3 ENST00000417377.1 |
HBD |
hemoglobin, delta |
| chr11_-_82708519 | 0.44 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr17_+_25799008 | 0.43 |
ENST00000583370.1 ENST00000398988.3 ENST00000268763.6 |
KSR1 |
kinase suppressor of ras 1 |
| chr5_-_141061777 | 0.42 |
ENST00000239440.4 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr19_+_13842559 | 0.41 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr17_-_7590745 | 0.40 |
ENST00000514944.1 ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53 |
tumor protein p53 |
| chr9_-_73029540 | 0.40 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr20_-_62129163 | 0.40 |
ENST00000298049.7 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr16_-_67281413 | 0.38 |
ENST00000258201.4 |
FHOD1 |
formin homology 2 domain containing 1 |
| chr16_+_19467772 | 0.37 |
ENST00000219821.5 ENST00000561503.1 ENST00000564959.1 |
TMC5 |
transmembrane channel-like 5 |
| chr18_+_55888767 | 0.37 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chrX_+_99839799 | 0.37 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr14_+_73525229 | 0.36 |
ENST00000527432.1 ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25 |
RNA binding motif protein 25 |
| chr2_-_152830441 | 0.36 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr3_-_11762202 | 0.36 |
ENST00000445411.1 ENST00000404339.1 ENST00000273038.3 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr12_-_7818474 | 0.35 |
ENST00000229304.4 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr6_+_63921351 | 0.34 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr14_+_58765103 | 0.34 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr4_-_122085469 | 0.33 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr14_+_73525144 | 0.33 |
ENST00000261973.7 ENST00000540173.1 |
RBM25 |
RNA binding motif protein 25 |
| chr13_-_31038370 | 0.33 |
ENST00000399489.1 ENST00000339872.4 |
HMGB1 |
high mobility group box 1 |
| chr20_+_48429233 | 0.32 |
ENST00000417961.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr15_-_74504597 | 0.32 |
ENST00000416286.3 |
STRA6 |
stimulated by retinoic acid 6 |
| chr2_+_201980827 | 0.32 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr10_-_81708854 | 0.30 |
ENST00000372292.3 |
SFTPD |
surfactant protein D |
| chr16_+_31366455 | 0.30 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr12_+_52695617 | 0.29 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr3_+_155860751 | 0.29 |
ENST00000471742.1 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr20_+_48429356 | 0.29 |
ENST00000361573.2 ENST00000541138.1 ENST00000539601.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr10_+_18549645 | 0.26 |
ENST00000396576.2 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_-_152830479 | 0.26 |
ENST00000360283.6 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_+_236687881 | 0.26 |
ENST00000526634.1 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr18_-_5419797 | 0.23 |
ENST00000542146.1 ENST00000427684.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr1_+_41204506 | 0.23 |
ENST00000525290.1 ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC |
nuclear transcription factor Y, gamma |
| chr8_+_128426535 | 0.21 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr10_-_14996017 | 0.19 |
ENST00000378241.1 ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr18_+_32290218 | 0.17 |
ENST00000348997.5 ENST00000588949.1 ENST00000597599.1 |
DTNA |
dystrobrevin, alpha |
| chr3_-_150996239 | 0.17 |
ENST00000309170.3 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
| chr14_-_25103472 | 0.16 |
ENST00000216341.4 ENST00000382542.1 ENST00000382540.1 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr1_-_117753540 | 0.16 |
ENST00000328189.3 ENST00000369458.3 |
VTCN1 |
V-set domain containing T cell activation inhibitor 1 |
| chr14_-_25103388 | 0.16 |
ENST00000526004.1 ENST00000415355.3 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr1_-_153513170 | 0.15 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr5_-_138533973 | 0.15 |
ENST00000507002.1 ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr17_+_30771279 | 0.15 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr10_-_14996070 | 0.13 |
ENST00000378258.1 ENST00000453695.2 ENST00000378246.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr1_+_117297007 | 0.13 |
ENST00000369478.3 ENST00000369477.1 |
CD2 |
CD2 molecule |
| chr9_-_127263265 | 0.12 |
ENST00000373587.3 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr11_-_4719072 | 0.12 |
ENST00000396950.3 ENST00000532598.1 |
OR51E2 |
olfactory receptor, family 51, subfamily E, member 2 |
| chr10_+_5005598 | 0.11 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr14_+_73525265 | 0.10 |
ENST00000525161.1 |
RBM25 |
RNA binding motif protein 25 |
| chr11_-_14913190 | 0.10 |
ENST00000532378.1 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr22_-_39715600 | 0.10 |
ENST00000427905.1 ENST00000402527.1 ENST00000216146.4 |
RPL3 |
ribosomal protein L3 |
| chr3_-_52713729 | 0.09 |
ENST00000296302.7 ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1 |
polybromo 1 |
| chr1_+_213224572 | 0.08 |
ENST00000543470.1 ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr3_-_87325728 | 0.05 |
ENST00000350375.2 |
POU1F1 |
POU class 1 homeobox 1 |
| chr19_+_11546093 | 0.05 |
ENST00000591462.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr16_-_72206034 | 0.05 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr13_+_109248500 | 0.05 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr3_-_46608010 | 0.05 |
ENST00000395905.3 |
LRRC2 |
leucine rich repeat containing 2 |
| chr11_+_59856130 | 0.03 |
ENST00000278888.3 |
MS4A2 |
membrane-spanning 4-domains, subfamily A, member 2 |
| chr19_-_54872556 | 0.02 |
ENST00000444687.1 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
| chr2_+_161993465 | 0.01 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr17_-_73150629 | 0.00 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.2 | 3.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.9 | 3.7 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.7 | 4.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.3 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.3 | 2.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 0.9 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.6 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 2.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0047023 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 3.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 2.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 1.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 6.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 7.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 7.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.2 | 3.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.7 | 4.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.5 | 3.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.4 | 1.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 7.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 1.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 1.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.9 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 2.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 2.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 1.7 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 1.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.9 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 2.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.7 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.9 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.4 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.3 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 4.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.3 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 1.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 2.0 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.3 | GO:1903944 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.5 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.9 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 3.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 2.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 1.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |