Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
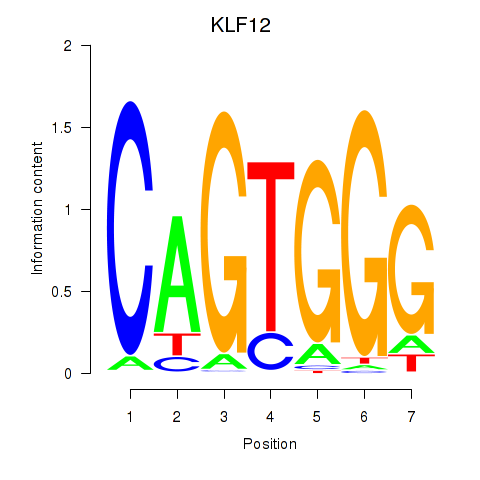
Results for KLF12
Z-value: 0.72
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | KLF12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | -0.22 | 4.1e-01 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_1665253 | 2.16 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_+_1665345 | 1.31 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr2_-_208634287 | 1.19 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chr15_+_89402148 | 1.10 |
ENST00000560601.1 |
ACAN |
aggrecan |
| chrX_+_152953505 | 1.03 |
ENST00000253122.5 |
SLC6A8 |
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr2_-_220435963 | 1.00 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr17_-_47841485 | 0.96 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr10_-_52645379 | 0.95 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr10_-_52645416 | 0.92 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr1_+_109792641 | 0.88 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr12_-_54691668 | 0.82 |
ENST00000553198.1 |
NFE2 |
nuclear factor, erythroid 2 |
| chrX_+_48644962 | 0.80 |
ENST00000376670.3 ENST00000376665.3 |
GATA1 |
GATA binding protein 1 (globin transcription factor 1) |
| chr1_+_116915855 | 0.79 |
ENST00000295598.5 |
ATP1A1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_+_214161854 | 0.78 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr2_-_192711968 | 0.77 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr3_-_49722523 | 0.74 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr5_+_137801160 | 0.74 |
ENST00000239938.4 |
EGR1 |
early growth response 1 |
| chr11_-_2160180 | 0.73 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr16_+_85646891 | 0.72 |
ENST00000393243.1 |
GSE1 |
Gse1 coiled-coil protein |
| chr6_+_31895467 | 0.72 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr3_-_8686479 | 0.71 |
ENST00000544814.1 ENST00000427408.1 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chr6_+_31895480 | 0.70 |
ENST00000418949.2 ENST00000383177.3 ENST00000477310.1 |
C2 CFB |
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr11_-_32456891 | 0.67 |
ENST00000452863.3 |
WT1 |
Wilms tumor 1 |
| chr10_+_21823079 | 0.66 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr15_+_84115868 | 0.58 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr14_+_32546274 | 0.58 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr3_+_193853927 | 0.58 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr16_+_85646763 | 0.56 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr19_-_55866061 | 0.56 |
ENST00000588572.2 ENST00000593184.1 ENST00000589467.1 |
COX6B2 |
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr1_+_214161272 | 0.55 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr17_+_27071002 | 0.54 |
ENST00000262395.5 ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4 |
TNF receptor-associated factor 4 |
| chr6_+_33168597 | 0.54 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr10_+_21823243 | 0.53 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr6_+_33168637 | 0.52 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr20_+_34742650 | 0.52 |
ENST00000373945.1 ENST00000338074.2 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr2_-_27593180 | 0.52 |
ENST00000493344.2 ENST00000445933.2 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr16_-_30798492 | 0.51 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chrX_-_118739835 | 0.49 |
ENST00000542113.1 ENST00000304449.5 |
NKRF |
NFKB repressing factor |
| chr17_-_8534067 | 0.48 |
ENST00000360416.3 ENST00000269243.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr12_-_71148413 | 0.48 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr17_-_8534031 | 0.47 |
ENST00000411957.1 ENST00000396239.1 ENST00000379980.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr2_+_128180842 | 0.47 |
ENST00000402125.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr2_-_111291587 | 0.47 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr1_+_24117693 | 0.46 |
ENST00000374503.3 ENST00000374502.3 |
LYPLA2 |
lysophospholipase II |
| chr3_-_52567792 | 0.45 |
ENST00000307092.4 ENST00000422318.2 ENST00000459839.1 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
| chr12_-_71148357 | 0.44 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr16_+_29819096 | 0.44 |
ENST00000568411.1 ENST00000563012.1 ENST00000562557.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_-_39330818 | 0.44 |
ENST00000594769.1 ENST00000602021.1 |
AC104534.3 |
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr19_+_10812108 | 0.44 |
ENST00000250237.5 ENST00000592254.1 |
QTRT1 |
queuine tRNA-ribosyltransferase 1 |
| chr1_+_114447763 | 0.44 |
ENST00000369563.3 |
DCLRE1B |
DNA cross-link repair 1B |
| chr16_+_29819372 | 0.43 |
ENST00000568544.1 ENST00000569978.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr13_-_67802549 | 0.43 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr4_+_128702969 | 0.43 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr16_+_29819446 | 0.42 |
ENST00000568282.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_32457075 | 0.40 |
ENST00000448076.3 |
WT1 |
Wilms tumor 1 |
| chr17_-_38978847 | 0.40 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr12_+_53848505 | 0.40 |
ENST00000552819.1 ENST00000455667.3 |
PCBP2 |
poly(rC) binding protein 2 |
| chr2_-_27593306 | 0.40 |
ENST00000347454.4 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr2_+_32288725 | 0.39 |
ENST00000315285.3 |
SPAST |
spastin |
| chr1_+_153600869 | 0.39 |
ENST00000292169.1 ENST00000368696.3 ENST00000436839.1 |
S100A1 |
S100 calcium binding protein A1 |
| chr1_+_24117627 | 0.39 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr7_+_143013198 | 0.39 |
ENST00000343257.2 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
| chr9_+_35673853 | 0.38 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr16_-_67978016 | 0.38 |
ENST00000264005.5 |
LCAT |
lecithin-cholesterol acyltransferase |
| chr17_+_79989937 | 0.38 |
ENST00000580965.1 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_-_212965104 | 0.37 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chr2_-_153573965 | 0.36 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr5_+_131705438 | 0.36 |
ENST00000245407.3 |
SLC22A5 |
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr1_-_114355083 | 0.35 |
ENST00000261441.5 |
RSBN1 |
round spermatid basic protein 1 |
| chr16_-_103572 | 0.35 |
ENST00000293860.5 |
POLR3K |
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr17_+_80477571 | 0.34 |
ENST00000335255.5 |
FOXK2 |
forkhead box K2 |
| chr20_+_34713312 | 0.33 |
ENST00000373946.3 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr7_-_135433460 | 0.33 |
ENST00000415751.1 |
FAM180A |
family with sequence similarity 180, member A |
| chr7_+_94537542 | 0.33 |
ENST00000433881.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_32288657 | 0.33 |
ENST00000345662.1 |
SPAST |
spastin |
| chr20_-_3154162 | 0.33 |
ENST00000360342.3 |
LZTS3 |
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr7_-_140178726 | 0.32 |
ENST00000480552.1 |
MKRN1 |
makorin ring finger protein 1 |
| chr16_-_20911641 | 0.32 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr1_-_19229014 | 0.32 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr17_-_41174424 | 0.32 |
ENST00000355653.3 |
VAT1 |
vesicle amine transport 1 |
| chr6_+_150070857 | 0.32 |
ENST00000544496.1 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_+_155178481 | 0.31 |
ENST00000368376.3 |
MTX1 |
metaxin 1 |
| chr1_-_202777535 | 0.31 |
ENST00000367264.2 |
KDM5B |
lysine (K)-specific demethylase 5B |
| chr4_+_128703295 | 0.31 |
ENST00000296464.4 ENST00000508549.1 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr5_-_32444828 | 0.31 |
ENST00000265069.8 |
ZFR |
zinc finger RNA binding protein |
| chr16_+_29818857 | 0.31 |
ENST00000567444.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_-_179227540 | 0.29 |
ENST00000520875.1 |
MGAT4B |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr6_+_150070831 | 0.28 |
ENST00000367380.5 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr14_-_65346555 | 0.28 |
ENST00000542895.1 ENST00000556626.1 |
SPTB |
spectrin, beta, erythrocytic |
| chr2_+_231902193 | 0.27 |
ENST00000373640.4 |
C2orf72 |
chromosome 2 open reading frame 72 |
| chr2_-_153573887 | 0.27 |
ENST00000493468.2 ENST00000545856.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_-_67142710 | 0.27 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr17_-_36906058 | 0.27 |
ENST00000580830.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr19_+_45312347 | 0.26 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr17_-_72864739 | 0.26 |
ENST00000579893.1 ENST00000544854.1 |
FDXR |
ferredoxin reductase |
| chr4_-_74904398 | 0.26 |
ENST00000296026.4 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
| chr10_-_81205373 | 0.25 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr16_+_28875126 | 0.25 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr7_-_79082867 | 0.25 |
ENST00000419488.1 ENST00000354212.4 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_+_12040238 | 0.25 |
ENST00000444836.1 ENST00000235329.5 |
MFN2 |
mitofusin 2 |
| chr11_-_61129335 | 0.25 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chrX_-_129299847 | 0.24 |
ENST00000319908.3 ENST00000287295.3 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr1_+_44870866 | 0.24 |
ENST00000355387.2 ENST00000361799.2 |
RNF220 |
ring finger protein 220 |
| chr3_+_148508845 | 0.24 |
ENST00000491148.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr1_+_155178518 | 0.24 |
ENST00000316721.4 |
MTX1 |
metaxin 1 |
| chr11_-_47400062 | 0.23 |
ENST00000533030.1 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr20_+_8112824 | 0.23 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr6_+_35995552 | 0.23 |
ENST00000468133.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr11_-_47399942 | 0.23 |
ENST00000227163.4 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr6_-_30640811 | 0.23 |
ENST00000376442.3 |
DHX16 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr6_+_27775899 | 0.22 |
ENST00000358739.3 |
HIST1H2AI |
histone cluster 1, H2ai |
| chr3_+_4535155 | 0.22 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr17_-_36891830 | 0.22 |
ENST00000578487.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr7_+_128502895 | 0.22 |
ENST00000492758.1 |
ATP6V1F |
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr6_-_114292449 | 0.22 |
ENST00000519065.1 |
HDAC2 |
histone deacetylase 2 |
| chr6_-_31864977 | 0.21 |
ENST00000395728.3 ENST00000375528.4 |
EHMT2 |
euchromatic histone-lysine N-methyltransferase 2 |
| chr10_-_51623203 | 0.21 |
ENST00000444743.1 ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23 |
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr11_-_46142948 | 0.21 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr2_+_99953816 | 0.21 |
ENST00000289371.6 |
EIF5B |
eukaryotic translation initiation factor 5B |
| chr17_+_7487146 | 0.21 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr3_-_133614421 | 0.21 |
ENST00000543906.1 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr11_-_47400078 | 0.21 |
ENST00000378538.3 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr16_+_87636474 | 0.21 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chr13_-_67804445 | 0.21 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr7_+_2394445 | 0.20 |
ENST00000360876.4 ENST00000413917.1 ENST00000397011.2 |
EIF3B |
eukaryotic translation initiation factor 3, subunit B |
| chr2_+_89952792 | 0.20 |
ENST00000390265.2 |
IGKV1D-33 |
immunoglobulin kappa variable 1D-33 |
| chr11_+_118889456 | 0.20 |
ENST00000528230.1 ENST00000525303.1 ENST00000434101.2 ENST00000359005.4 ENST00000533058.1 |
TRAPPC4 |
trafficking protein particle complex 4 |
| chr1_+_150229554 | 0.20 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr15_+_64428529 | 0.20 |
ENST00000560861.1 |
SNX1 |
sorting nexin 1 |
| chr10_-_103880209 | 0.20 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr11_+_20620946 | 0.19 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr1_-_43205811 | 0.19 |
ENST00000372539.3 ENST00000296387.1 ENST00000539749.1 |
CLDN19 |
claudin 19 |
| chr3_-_133614597 | 0.19 |
ENST00000285208.4 ENST00000460865.3 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr11_+_125365110 | 0.19 |
ENST00000527818.1 |
AP000708.1 |
AP000708.1 |
| chr5_+_137688285 | 0.19 |
ENST00000314358.5 |
KDM3B |
lysine (K)-specific demethylase 3B |
| chr17_+_4618734 | 0.19 |
ENST00000571206.1 |
ARRB2 |
arrestin, beta 2 |
| chr5_+_139493665 | 0.19 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr1_-_31538517 | 0.19 |
ENST00000440538.2 ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1 |
pumilio RNA-binding family member 1 |
| chr6_-_27775694 | 0.19 |
ENST00000377401.2 |
HIST1H2BL |
histone cluster 1, H2bl |
| chr22_+_47016277 | 0.19 |
ENST00000406902.1 |
GRAMD4 |
GRAM domain containing 4 |
| chr3_-_148804275 | 0.18 |
ENST00000392912.2 ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF |
helicase-like transcription factor |
| chr1_-_32403903 | 0.18 |
ENST00000344035.6 ENST00000356536.3 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr13_-_52027134 | 0.18 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr15_-_34394119 | 0.18 |
ENST00000256545.4 |
EMC7 |
ER membrane protein complex subunit 7 |
| chr19_+_6772710 | 0.18 |
ENST00000304076.2 ENST00000602142.1 ENST00000596764.1 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
| chr2_-_89568263 | 0.18 |
ENST00000473726.1 |
IGKV1-33 |
immunoglobulin kappa variable 1-33 |
| chr6_-_114292284 | 0.17 |
ENST00000520895.1 ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2 |
histone deacetylase 2 |
| chr16_+_770975 | 0.17 |
ENST00000569529.1 ENST00000564000.1 ENST00000219535.3 |
FAM173A |
family with sequence similarity 173, member A |
| chr16_+_67876180 | 0.17 |
ENST00000303596.1 |
THAP11 |
THAP domain containing 11 |
| chr17_-_47045949 | 0.17 |
ENST00000357424.2 |
GIP |
gastric inhibitory polypeptide |
| chr4_-_168155417 | 0.17 |
ENST00000511269.1 ENST00000506697.1 ENST00000512042.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_-_140178775 | 0.17 |
ENST00000474576.1 ENST00000473444.1 ENST00000471104.1 |
MKRN1 |
makorin ring finger protein 1 |
| chr12_+_120740119 | 0.17 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr3_+_184037466 | 0.17 |
ENST00000441154.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr8_+_124194752 | 0.17 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr7_+_140373619 | 0.17 |
ENST00000483369.1 |
ADCK2 |
aarF domain containing kinase 2 |
| chr6_+_35995531 | 0.17 |
ENST00000229794.4 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr11_-_64013288 | 0.16 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr12_+_118573663 | 0.16 |
ENST00000261313.2 |
PEBP1 |
phosphatidylethanolamine binding protein 1 |
| chr6_+_108487245 | 0.16 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr8_+_75736761 | 0.16 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr6_-_43021612 | 0.16 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
| chr1_+_6845384 | 0.15 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr12_+_53848549 | 0.15 |
ENST00000439930.3 ENST00000548933.1 ENST00000562264.1 |
PCBP2 |
poly(rC) binding protein 2 |
| chr1_+_156084461 | 0.15 |
ENST00000347559.2 ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA |
lamin A/C |
| chr14_+_102276209 | 0.15 |
ENST00000445439.3 ENST00000334743.5 ENST00000557095.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr8_+_22022800 | 0.15 |
ENST00000397814.3 |
BMP1 |
bone morphogenetic protein 1 |
| chr14_-_74417096 | 0.15 |
ENST00000286544.3 |
FAM161B |
family with sequence similarity 161, member B |
| chr1_-_216596738 | 0.15 |
ENST00000307340.3 ENST00000366943.2 ENST00000366942.3 |
USH2A |
Usher syndrome 2A (autosomal recessive, mild) |
| chr3_-_45837959 | 0.15 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr1_-_244615425 | 0.15 |
ENST00000366535.3 |
ADSS |
adenylosuccinate synthase |
| chrX_-_54209640 | 0.15 |
ENST00000375180.2 ENST00000328235.4 ENST00000477084.1 |
FAM120C |
family with sequence similarity 120C |
| chr14_-_75530693 | 0.15 |
ENST00000555135.1 ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr17_-_27418537 | 0.14 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_+_16083154 | 0.14 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr13_-_46716969 | 0.14 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr10_-_115933942 | 0.14 |
ENST00000369285.3 ENST00000369287.3 ENST00000369286.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr1_+_226736446 | 0.14 |
ENST00000366788.3 ENST00000366789.4 |
C1orf95 |
chromosome 1 open reading frame 95 |
| chr17_+_26989109 | 0.14 |
ENST00000314616.6 ENST00000347486.4 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr10_-_65028938 | 0.14 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chrX_-_129299638 | 0.14 |
ENST00000535724.1 ENST00000346424.2 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr10_-_65028817 | 0.14 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr8_-_38386175 | 0.14 |
ENST00000437935.2 ENST00000358138.1 |
C8orf86 |
chromosome 8 open reading frame 86 |
| chr7_-_38403077 | 0.14 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr17_+_7155343 | 0.14 |
ENST00000573513.1 ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr6_+_31730773 | 0.13 |
ENST00000415669.2 ENST00000425424.1 |
SAPCD1 |
suppressor APC domain containing 1 |
| chr16_+_103816 | 0.13 |
ENST00000383018.3 ENST00000417493.1 |
SNRNP25 |
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr15_-_34394008 | 0.13 |
ENST00000527822.1 ENST00000528949.1 |
EMC7 |
ER membrane protein complex subunit 7 |
| chr20_-_34287259 | 0.13 |
ENST00000397425.1 ENST00000540053.1 ENST00000541387.1 ENST00000374092.4 |
NFS1 |
NFS1 cysteine desulfurase |
| chr14_-_23772032 | 0.13 |
ENST00000452015.4 |
PPP1R3E |
protein phosphatase 1, regulatory subunit 3E |
| chr2_-_50201327 | 0.13 |
ENST00000412315.1 |
NRXN1 |
neurexin 1 |
| chr9_+_131445928 | 0.13 |
ENST00000372692.4 |
SET |
SET nuclear oncogene |
| chr9_-_94877658 | 0.13 |
ENST00000262554.2 ENST00000337841.4 |
SPTLC1 |
serine palmitoyltransferase, long chain base subunit 1 |
| chr12_-_82153087 | 0.13 |
ENST00000547623.1 ENST00000549396.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr19_-_46272106 | 0.13 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chr4_-_116034979 | 0.13 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr2_-_225266711 | 0.13 |
ENST00000389874.3 |
FAM124B |
family with sequence similarity 124B |
| chr6_-_33160231 | 0.13 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chr15_-_43877062 | 0.13 |
ENST00000381885.1 ENST00000396923.3 |
PPIP5K1 |
diphosphoinositol pentakisphosphate kinase 1 |
| chr17_+_57784826 | 0.13 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.4 | 1.3 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.4 | 1.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.4 | 1.8 | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) |
| 0.3 | 1.0 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.3 | 0.3 | GO:0010637 | regulation of mitochondrial fusion(GO:0010635) negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 0.6 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 | 0.8 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.2 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 0.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 0.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.9 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 0.9 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.8 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.1 | 0.3 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.4 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.7 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.4 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.1 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.4 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.4 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.2 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0045116 | response to UV-C(GO:0010225) protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 3.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.3 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.9 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0000995 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |