Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
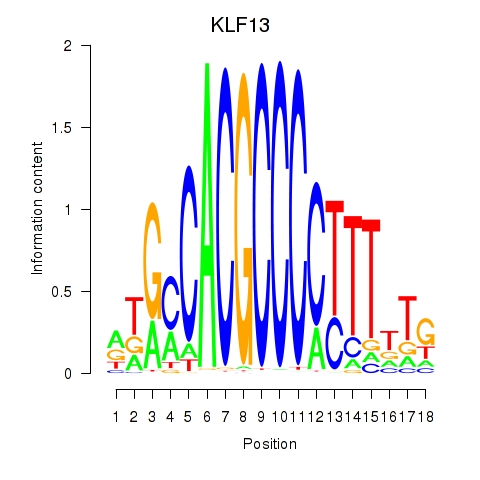
Results for KLF13
Z-value: 0.94
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | KLF13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF13 | hg19_v2_chr15_+_31619013_31619095 | -0.15 | 5.8e-01 | Click! |
Activity profile of KLF13 motif
Sorted Z-values of KLF13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_882966 | 3.08 |
ENST00000336868.3 |
NXN |
nucleoredoxin |
| chrX_-_140271249 | 2.60 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr19_+_45349432 | 2.22 |
ENST00000252485.4 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr6_-_31697977 | 1.68 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_31697563 | 1.41 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr7_-_111846435 | 1.37 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr6_-_2962331 | 1.24 |
ENST00000380524.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr13_+_27131798 | 1.05 |
ENST00000361042.4 |
WASF3 |
WAS protein family, member 3 |
| chr11_+_129939779 | 1.04 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr11_+_129939811 | 1.02 |
ENST00000345598.5 ENST00000338167.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr11_-_82612727 | 0.99 |
ENST00000531128.1 ENST00000535099.1 ENST00000527444.1 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
| chr3_+_132136331 | 0.91 |
ENST00000260818.6 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr2_-_110371777 | 0.90 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr4_+_55524085 | 0.90 |
ENST00000412167.2 ENST00000288135.5 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr2_-_110371720 | 0.88 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr2_+_71295733 | 0.86 |
ENST00000443938.2 ENST00000244204.6 |
NAGK |
N-acetylglucosamine kinase |
| chr11_+_123986069 | 0.86 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr8_-_145013711 | 0.85 |
ENST00000345136.3 |
PLEC |
plectin |
| chr2_-_110371412 | 0.85 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr14_+_93260642 | 0.83 |
ENST00000355976.2 |
GOLGA5 |
golgin A5 |
| chr14_+_24867992 | 0.83 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr19_+_54695098 | 0.82 |
ENST00000396388.2 |
TSEN34 |
TSEN34 tRNA splicing endonuclease subunit |
| chr12_-_124457257 | 0.81 |
ENST00000545891.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr17_-_19281203 | 0.80 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr8_-_62627057 | 0.79 |
ENST00000519234.1 ENST00000379449.6 ENST00000379454.4 ENST00000518068.1 ENST00000517856.1 ENST00000356457.5 |
ASPH |
aspartate beta-hydroxylase |
| chr5_+_148206156 | 0.78 |
ENST00000305988.4 |
ADRB2 |
adrenoceptor beta 2, surface |
| chr13_+_27131887 | 0.77 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chr12_-_124457371 | 0.74 |
ENST00000238156.3 ENST00000545037.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chrX_+_54835493 | 0.72 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr1_-_16939976 | 0.68 |
ENST00000430580.2 |
NBPF1 |
neuroblastoma breakpoint family, member 1 |
| chr20_-_44539538 | 0.67 |
ENST00000372420.1 |
PLTP |
phospholipid transfer protein |
| chr6_-_31697255 | 0.66 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_3074231 | 0.65 |
ENST00000572355.1 ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr16_-_3073933 | 0.65 |
ENST00000574151.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr2_+_71295717 | 0.63 |
ENST00000418807.3 ENST00000443872.2 |
NAGK |
N-acetylglucosamine kinase |
| chr1_-_146082633 | 0.62 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr4_+_56719782 | 0.60 |
ENST00000381295.2 ENST00000346134.7 ENST00000349598.6 |
EXOC1 |
exocyst complex component 1 |
| chr10_-_13344341 | 0.60 |
ENST00000396920.3 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
| chr3_+_50606901 | 0.58 |
ENST00000455834.1 |
HEMK1 |
HemK methyltransferase family member 1 |
| chr17_-_7165662 | 0.57 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr6_-_159065741 | 0.55 |
ENST00000367085.3 ENST00000367089.3 |
DYNLT1 |
dynein, light chain, Tctex-type 1 |
| chr19_-_15236562 | 0.54 |
ENST00000263383.3 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr3_+_184529948 | 0.54 |
ENST00000436792.2 ENST00000446204.2 ENST00000422105.1 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_+_50606577 | 0.52 |
ENST00000434410.1 ENST00000232854.4 |
HEMK1 |
HemK methyltransferase family member 1 |
| chr19_+_41284121 | 0.51 |
ENST00000594800.1 ENST00000357052.2 ENST00000602173.1 |
RAB4B |
RAB4B, member RAS oncogene family |
| chr2_-_110371664 | 0.50 |
ENST00000545389.1 ENST00000423520.1 |
SEPT10 |
septin 10 |
| chr6_-_33285505 | 0.49 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr19_-_12886327 | 0.48 |
ENST00000397668.3 ENST00000587178.1 ENST00000264827.5 |
HOOK2 |
hook microtubule-tethering protein 2 |
| chr1_+_21766641 | 0.48 |
ENST00000342104.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr3_+_184529929 | 0.48 |
ENST00000287546.4 ENST00000437079.3 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr17_+_19281034 | 0.47 |
ENST00000308406.5 ENST00000299612.7 |
MAPK7 |
mitogen-activated protein kinase 7 |
| chr6_+_52535878 | 0.46 |
ENST00000211314.4 |
TMEM14A |
transmembrane protein 14A |
| chr15_+_23255242 | 0.46 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr2_-_85645545 | 0.46 |
ENST00000409275.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr14_-_74551172 | 0.45 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr15_-_75017711 | 0.45 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr1_+_21766588 | 0.45 |
ENST00000454000.2 ENST00000318220.6 ENST00000318249.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr10_+_134351319 | 0.45 |
ENST00000368594.3 ENST00000368593.3 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
| chr2_-_209119831 | 0.44 |
ENST00000345146.2 |
IDH1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr6_+_31588478 | 0.44 |
ENST00000376007.4 ENST00000376033.2 |
PRRC2A |
proline-rich coiled-coil 2A |
| chr14_+_93260569 | 0.42 |
ENST00000163416.2 |
GOLGA5 |
golgin A5 |
| chr7_+_100210133 | 0.40 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr16_+_84853580 | 0.40 |
ENST00000262424.5 ENST00000566151.1 ENST00000567845.1 ENST00000564567.1 ENST00000569090.1 |
CRISPLD2 |
cysteine-rich secretory protein LCCL domain containing 2 |
| chr11_+_121163466 | 0.39 |
ENST00000527762.1 ENST00000534230.1 ENST00000392789.2 |
SC5D |
sterol-C5-desaturase |
| chr1_+_42922173 | 0.39 |
ENST00000455780.1 ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr19_+_49713991 | 0.37 |
ENST00000597316.1 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_39616410 | 0.36 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr7_+_100209725 | 0.35 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr17_+_42264322 | 0.35 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr7_+_100209979 | 0.33 |
ENST00000493970.1 ENST00000379527.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr12_+_121124921 | 0.33 |
ENST00000412616.2 |
MLEC |
malectin |
| chr7_+_33169142 | 0.32 |
ENST00000242067.6 ENST00000350941.3 ENST00000396127.2 ENST00000355070.2 ENST00000354265.4 ENST00000425508.2 |
BBS9 |
Bardet-Biedl syndrome 9 |
| chr7_+_12727250 | 0.32 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr10_-_111683308 | 0.32 |
ENST00000502935.1 ENST00000322238.8 ENST00000369680.4 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr16_+_4526341 | 0.31 |
ENST00000458134.3 ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2 |
heme oxygenase (decycling) 2 |
| chr5_+_170288856 | 0.31 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr3_+_158519654 | 0.30 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr7_+_100271355 | 0.30 |
ENST00000436220.1 ENST00000424361.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr11_-_6426635 | 0.30 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chrX_-_103401649 | 0.30 |
ENST00000357421.4 |
SLC25A53 |
solute carrier family 25, member 53 |
| chr14_-_74551096 | 0.29 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr12_-_113772835 | 0.29 |
ENST00000552014.1 ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr7_+_100271446 | 0.28 |
ENST00000419828.1 ENST00000427895.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr15_-_30685752 | 0.28 |
ENST00000299847.2 ENST00000397827.3 |
CHRFAM7A |
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr7_+_100450328 | 0.27 |
ENST00000540482.1 ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9 |
solute carrier family 12, member 9 |
| chr1_+_42921761 | 0.27 |
ENST00000372562.1 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr14_+_75469606 | 0.27 |
ENST00000266126.5 |
EIF2B2 |
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr16_-_28936493 | 0.27 |
ENST00000544477.1 ENST00000357573.6 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr19_-_50370509 | 0.26 |
ENST00000596014.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr4_+_95373037 | 0.26 |
ENST00000359265.4 ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5 |
PDZ and LIM domain 5 |
| chr17_+_42264395 | 0.25 |
ENST00000587989.1 ENST00000590235.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr7_-_99756293 | 0.25 |
ENST00000316937.3 ENST00000456769.1 |
C7orf43 |
chromosome 7 open reading frame 43 |
| chr17_-_40333150 | 0.25 |
ENST00000264661.3 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr1_+_172502336 | 0.25 |
ENST00000263688.3 |
SUCO |
SUN domain containing ossification factor |
| chr11_-_27494279 | 0.24 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr19_-_14629224 | 0.24 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr19_-_15236470 | 0.24 |
ENST00000533747.1 ENST00000598709.1 ENST00000534378.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr20_+_48599506 | 0.24 |
ENST00000244050.2 |
SNAI1 |
snail family zinc finger 1 |
| chr17_+_27055798 | 0.23 |
ENST00000268766.6 |
NEK8 |
NIMA-related kinase 8 |
| chr21_+_48055527 | 0.23 |
ENST00000397638.2 ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2 |
protein arginine methyltransferase 2 |
| chr19_+_36631867 | 0.23 |
ENST00000588780.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr15_+_84904525 | 0.23 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr11_-_27494309 | 0.23 |
ENST00000389858.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr12_-_48152853 | 0.22 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr12_+_56521840 | 0.21 |
ENST00000394048.5 |
ESYT1 |
extended synaptotagmin-like protein 1 |
| chr19_+_36236514 | 0.21 |
ENST00000222266.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr17_-_5487768 | 0.21 |
ENST00000269280.4 ENST00000345221.3 ENST00000262467.5 |
NLRP1 |
NLR family, pyrin domain containing 1 |
| chr19_+_36632056 | 0.21 |
ENST00000586851.1 ENST00000590211.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr12_+_58013693 | 0.20 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr20_+_34129770 | 0.20 |
ENST00000348547.2 ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3 |
ERGIC and golgi 3 |
| chr6_+_30594619 | 0.19 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr17_+_42264556 | 0.18 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr10_+_102672712 | 0.18 |
ENST00000370271.3 ENST00000370269.3 ENST00000609386.1 |
FAM178A |
family with sequence similarity 178, member A |
| chr15_+_83098710 | 0.17 |
ENST00000561062.1 ENST00000358583.3 |
GOLGA6L9 |
golgin A6 family-like 20 |
| chr10_-_111683183 | 0.17 |
ENST00000403138.2 ENST00000369683.1 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr9_-_35111570 | 0.17 |
ENST00000378561.1 ENST00000603301.1 |
FAM214B |
family with sequence similarity 214, member B |
| chr12_-_6580094 | 0.16 |
ENST00000361716.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr15_+_82722225 | 0.16 |
ENST00000300515.8 |
GOLGA6L9 |
golgin A6 family-like 9 |
| chr12_-_29534074 | 0.15 |
ENST00000546839.1 ENST00000360150.4 ENST00000552155.1 ENST00000550353.1 ENST00000548441.1 ENST00000552132.1 |
ERGIC2 |
ERGIC and golgi 2 |
| chr14_+_23067146 | 0.15 |
ENST00000428304.2 |
ABHD4 |
abhydrolase domain containing 4 |
| chr11_-_7041466 | 0.15 |
ENST00000536068.1 ENST00000278314.4 |
ZNF214 |
zinc finger protein 214 |
| chrX_+_47077632 | 0.15 |
ENST00000457458.2 |
CDK16 |
cyclin-dependent kinase 16 |
| chr13_-_30424821 | 0.15 |
ENST00000380680.4 |
UBL3 |
ubiquitin-like 3 |
| chr14_+_23299088 | 0.14 |
ENST00000355151.5 ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52 |
mitochondrial ribosomal protein L52 |
| chr19_+_49866851 | 0.13 |
ENST00000221498.2 ENST00000596402.1 |
DKKL1 |
dickkopf-like 1 |
| chr4_+_699610 | 0.13 |
ENST00000521023.2 |
PCGF3 |
polycomb group ring finger 3 |
| chr17_-_74497432 | 0.12 |
ENST00000590288.1 ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
| chr9_-_35111420 | 0.12 |
ENST00000378557.1 |
FAM214B |
family with sequence similarity 214, member B |
| chr13_+_100153665 | 0.12 |
ENST00000376387.4 |
TM9SF2 |
transmembrane 9 superfamily member 2 |
| chr8_+_26149007 | 0.11 |
ENST00000380737.3 ENST00000524169.1 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr17_-_77770830 | 0.11 |
ENST00000269385.4 |
CBX8 |
chromobox homolog 8 |
| chr17_-_5487277 | 0.11 |
ENST00000572272.1 ENST00000354411.3 ENST00000577119.1 |
NLRP1 |
NLR family, pyrin domain containing 1 |
| chr7_-_91763822 | 0.11 |
ENST00000003100.8 |
CYP51A1 |
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr19_-_10764509 | 0.11 |
ENST00000591501.1 |
ILF3-AS1 |
ILF3 antisense RNA 1 (head to head) |
| chr19_-_2151523 | 0.11 |
ENST00000350812.6 ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1 |
adaptor-related protein complex 3, delta 1 subunit |
| chr4_+_699537 | 0.11 |
ENST00000419774.1 ENST00000362003.5 ENST00000400151.2 ENST00000427463.1 ENST00000470161.2 |
PCGF3 |
polycomb group ring finger 3 |
| chr19_+_7985198 | 0.11 |
ENST00000221573.6 ENST00000595637.1 |
SNAPC2 |
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr17_+_74733744 | 0.10 |
ENST00000586689.1 ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11 |
major facilitator superfamily domain containing 11 |
| chr19_-_47164386 | 0.10 |
ENST00000391916.2 ENST00000410105.2 |
DACT3 |
dishevelled-binding antagonist of beta-catenin 3 |
| chr9_+_35605274 | 0.10 |
ENST00000336395.5 |
TESK1 |
testis-specific kinase 1 |
| chr12_-_6579808 | 0.10 |
ENST00000535180.1 ENST00000400911.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr10_+_104678102 | 0.10 |
ENST00000433628.2 |
CNNM2 |
cyclin M2 |
| chr17_+_73008755 | 0.10 |
ENST00000584208.1 ENST00000301585.5 |
ICT1 |
immature colon carcinoma transcript 1 |
| chr2_+_239335449 | 0.09 |
ENST00000264607.4 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
| chr11_-_89540388 | 0.09 |
ENST00000532501.2 |
TRIM49 |
tripartite motif containing 49 |
| chr7_-_91764108 | 0.09 |
ENST00000450723.1 |
CYP51A1 |
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr10_+_104005272 | 0.09 |
ENST00000369983.3 |
GBF1 |
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr20_+_37590942 | 0.09 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr6_+_28109703 | 0.09 |
ENST00000457389.2 ENST00000330236.6 |
ZKSCAN8 |
zinc finger with KRAB and SCAN domains 8 |
| chr19_+_36603662 | 0.09 |
ENST00000586670.1 |
OVOL3 |
ovo-like zinc finger 3 |
| chr16_+_4897912 | 0.08 |
ENST00000545171.1 |
UBN1 |
ubinuclein 1 |
| chr6_-_11779403 | 0.08 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr2_+_27994567 | 0.08 |
ENST00000379666.3 ENST00000296102.3 |
MRPL33 |
mitochondrial ribosomal protein L33 |
| chr19_+_36236491 | 0.08 |
ENST00000591949.1 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr6_-_43596899 | 0.08 |
ENST00000307126.5 ENST00000452781.1 |
GTPBP2 |
GTP binding protein 2 |
| chr15_-_26874230 | 0.08 |
ENST00000400188.3 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr20_+_60813535 | 0.08 |
ENST00000358053.2 ENST00000313733.3 ENST00000439951.2 |
OSBPL2 |
oxysterol binding protein-like 2 |
| chr2_+_201676256 | 0.08 |
ENST00000452206.1 ENST00000410110.2 ENST00000409600.1 |
BZW1 |
basic leucine zipper and W2 domains 1 |
| chr7_+_155089486 | 0.08 |
ENST00000340368.4 ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1 |
insulin induced gene 1 |
| chr20_+_3776371 | 0.07 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr6_+_33378738 | 0.07 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr1_-_25558963 | 0.07 |
ENST00000354361.3 |
SYF2 |
SYF2 pre-mRNA-splicing factor |
| chr2_-_97405775 | 0.06 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr15_-_74374891 | 0.06 |
ENST00000290438.3 |
GOLGA6A |
golgin A6 family, member A |
| chr10_+_104678032 | 0.06 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr18_-_51750948 | 0.05 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr13_+_45694583 | 0.05 |
ENST00000340473.6 |
GTF2F2 |
general transcription factor IIF, polypeptide 2, 30kDa |
| chr19_+_45458503 | 0.05 |
ENST00000337392.5 ENST00000591304.1 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
| chr6_-_11779014 | 0.05 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr9_-_70490107 | 0.05 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr1_-_25558984 | 0.05 |
ENST00000236273.4 |
SYF2 |
SYF2 pre-mRNA-splicing factor |
| chr9_-_131038266 | 0.05 |
ENST00000490628.1 ENST00000421699.2 ENST00000450617.1 |
GOLGA2 |
golgin A2 |
| chr11_+_49050504 | 0.04 |
ENST00000332682.7 |
TRIM49B |
tripartite motif containing 49B |
| chr7_+_99102573 | 0.04 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr6_-_31125850 | 0.03 |
ENST00000507751.1 ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1 |
coiled-coil alpha-helical rod protein 1 |
| chr2_-_73053126 | 0.03 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr6_-_11779174 | 0.03 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr20_-_44600810 | 0.03 |
ENST00000322927.2 ENST00000426788.1 |
ZNF335 |
zinc finger protein 335 |
| chr16_-_30905263 | 0.02 |
ENST00000572628.1 |
BCL7C |
B-cell CLL/lymphoma 7C |
| chr11_-_59383617 | 0.02 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr20_-_8000426 | 0.02 |
ENST00000527925.1 ENST00000246024.2 |
TMX4 |
thioredoxin-related transmembrane protein 4 |
| chr19_+_5455421 | 0.02 |
ENST00000222033.4 |
ZNRF4 |
zinc and ring finger 4 |
| chr17_-_13505219 | 0.02 |
ENST00000284110.1 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr19_+_45504688 | 0.02 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_-_8977714 | 0.02 |
ENST00000319688.5 ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220 |
kinase D-interacting substrate, 220kDa |
| chr12_+_121124599 | 0.01 |
ENST00000228506.3 |
MLEC |
malectin |
| chr19_+_44100632 | 0.01 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.6 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 2.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 1.5 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.4 | 3.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 0.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.4 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0042731 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.3 | 0.9 | GO:0048170 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 0.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 1.4 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.3 | 0.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.0 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.2 | 3.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.2 | 3.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.9 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.2 | 0.8 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 0.7 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.2 | 0.7 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.2 | 0.6 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.5 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.7 | GO:0033121 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:1903949 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.6 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.4 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 3.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |