Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
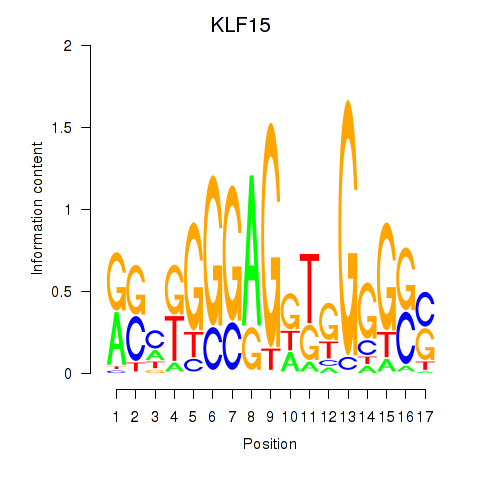
Results for KLF15
Z-value: 0.81
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | KLF15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | 0.16 | 5.5e-01 | Click! |
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
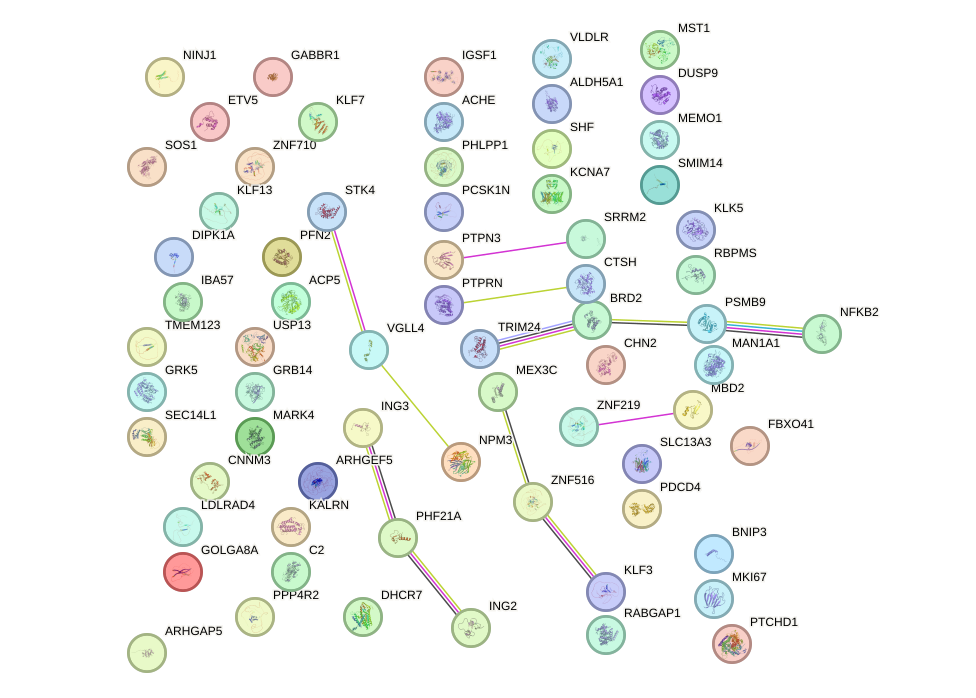
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29595779 | 2.82 |
ENST00000355973.3 ENST00000377012.4 |
GABBR1 |
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_94285637 | 2.22 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr14_+_32546274 | 1.84 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr2_-_20425158 | 1.71 |
ENST00000381150.1 |
SDC1 |
syndecan 1 |
| chr5_-_73937244 | 1.58 |
ENST00000302351.4 ENST00000510316.1 ENST00000508331.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr20_-_22565101 | 1.56 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr2_-_20424844 | 1.52 |
ENST00000403076.1 ENST00000254351.4 |
SDC1 |
syndecan 1 |
| chrX_+_152907913 | 1.12 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chrX_-_130423386 | 1.06 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chrX_-_130423240 | 1.01 |
ENST00000370910.1 ENST00000370901.4 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr6_+_31865552 | 0.99 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr9_-_95896550 | 0.95 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr14_+_32546485 | 0.93 |
ENST00000345122.3 ENST00000432921.1 ENST00000433497.1 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr18_-_74207146 | 0.93 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr15_-_79237433 | 0.91 |
ENST00000220166.5 |
CTSH |
cathepsin H |
| chr18_+_56338618 | 0.90 |
ENST00000348428.3 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chrX_+_154114635 | 0.88 |
ENST00000369446.2 |
F8A1 |
coagulation factor VIII-associated 1 |
| chrX_+_23352133 | 0.87 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr14_+_59655369 | 0.86 |
ENST00000360909.3 ENST00000351081.1 ENST00000556135.1 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
| chr8_+_30241934 | 0.81 |
ENST00000538486.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr20_-_33413416 | 0.81 |
ENST00000359003.2 |
NCOA6 |
nuclear receptor coactivator 6 |
| chr7_-_94285511 | 0.81 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr3_-_49726486 | 0.79 |
ENST00000449682.2 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chrX_-_48693955 | 0.74 |
ENST00000218230.5 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr7_-_94285472 | 0.72 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr9_-_112260531 | 0.70 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr2_-_73511559 | 0.70 |
ENST00000521871.1 |
FBXO41 |
F-box protein 41 |
| chr15_-_45480153 | 0.69 |
ENST00000560471.1 ENST00000560540.1 |
SHF |
Src homology 2 domain containing F |
| chrX_-_130423200 | 0.69 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr7_+_29519486 | 0.68 |
ENST00000409041.4 |
CHN2 |
chimerin 2 |
| chr7_-_94285402 | 0.67 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr17_+_48503519 | 0.67 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr1_-_93426998 | 0.67 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr10_+_112631547 | 0.66 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr14_-_89883412 | 0.64 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr2_-_242254595 | 0.63 |
ENST00000441124.1 ENST00000391976.2 |
HDLBP |
high density lipoprotein binding protein |
| chr16_+_2802623 | 0.62 |
ENST00000576924.1 ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr15_-_34875771 | 0.62 |
ENST00000267731.7 |
GOLGA8B |
golgin A8 family, member B |
| chr9_+_2622085 | 0.61 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr17_+_48503603 | 0.60 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr1_-_19229248 | 0.60 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr8_+_30241995 | 0.60 |
ENST00000397323.4 ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS |
RNA binding protein with multiple splicing |
| chr19_-_11689752 | 0.59 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr3_+_179370517 | 0.59 |
ENST00000263966.3 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr13_+_98795505 | 0.58 |
ENST00000319562.6 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr13_+_98795434 | 0.57 |
ENST00000376586.2 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr2_+_97481974 | 0.56 |
ENST00000377060.3 ENST00000305510.3 |
CNNM3 |
cyclin M3 |
| chr11_-_46142615 | 0.55 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr3_-_197282821 | 0.55 |
ENST00000445160.2 ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr20_-_45280066 | 0.55 |
ENST00000279027.4 |
SLC13A3 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr20_-_45280091 | 0.54 |
ENST00000396360.1 ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr3_-_11610255 | 0.54 |
ENST00000424529.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr12_-_31744031 | 0.53 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr9_+_2621798 | 0.52 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr7_+_65338230 | 0.52 |
ENST00000360768.3 |
VKORC1L1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr10_+_114709999 | 0.51 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_+_138145076 | 0.51 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr4_-_39640700 | 0.50 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr18_+_60382672 | 0.50 |
ENST00000400316.4 ENST00000262719.5 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
| chr3_-_149688502 | 0.50 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr10_-_133795416 | 0.50 |
ENST00000540159.1 ENST00000368636.4 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr18_+_13218769 | 0.50 |
ENST00000399848.3 ENST00000361205.4 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr2_+_201171242 | 0.49 |
ENST00000360760.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr6_+_6588316 | 0.49 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr1_+_228353495 | 0.49 |
ENST00000366711.3 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr6_-_119670919 | 0.48 |
ENST00000368468.3 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
| chr19_-_49576198 | 0.47 |
ENST00000221444.1 |
KCNA7 |
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr6_+_32811885 | 0.47 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr15_+_31619013 | 0.46 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr2_-_208030647 | 0.45 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr9_+_125703282 | 0.43 |
ENST00000373647.4 ENST00000402311.1 |
RABGAP1 |
RAB GTPase activating protein 1 |
| chr3_+_123813543 | 0.43 |
ENST00000360013.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr19_-_55866061 | 0.43 |
ENST00000588572.2 ENST00000593184.1 ENST00000589467.1 |
COX6B2 |
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr2_+_201171372 | 0.42 |
ENST00000409140.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr11_-_102323740 | 0.42 |
ENST00000398136.2 |
TMEM123 |
transmembrane protein 123 |
| chr6_+_32936353 | 0.42 |
ENST00000374825.4 |
BRD2 |
bromodomain containing 2 |
| chr10_-_129924611 | 0.41 |
ENST00000368654.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr11_+_3876859 | 0.41 |
ENST00000300737.4 |
STIM1 |
stromal interaction molecule 1 |
| chr17_+_75137034 | 0.41 |
ENST00000436233.4 ENST00000443798.4 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr10_+_120967072 | 0.41 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr19_+_45754505 | 0.40 |
ENST00000262891.4 ENST00000300843.4 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
| chr18_-_57364588 | 0.40 |
ENST00000439986.4 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
| chr14_-_21566731 | 0.39 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr3_+_73045936 | 0.39 |
ENST00000356692.5 ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2 |
protein phosphatase 4, regulatory subunit 2 |
| chr2_-_242255117 | 0.38 |
ENST00000420451.1 ENST00000417540.1 ENST00000310931.4 |
HDLBP |
high density lipoprotein binding protein |
| chr11_+_82783097 | 0.37 |
ENST00000501011.2 ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1 |
RAB30 antisense RNA 1 (head to head) |
| chr18_-_51750948 | 0.36 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr3_-_185826286 | 0.36 |
ENST00000537818.1 ENST00000422039.1 ENST00000434744.1 |
ETV5 |
ets variant 5 |
| chr18_-_48723690 | 0.36 |
ENST00000406189.3 |
MEX3C |
mex-3 RNA binding family member C |
| chr4_+_184426147 | 0.35 |
ENST00000302327.3 |
ING2 |
inhibitor of growth family, member 2 |
| chr19_-_51456344 | 0.35 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr4_+_38665810 | 0.35 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr15_+_90544532 | 0.34 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chr7_+_144052381 | 0.34 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr1_-_113498943 | 0.34 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr7_-_100493744 | 0.34 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr10_+_104155450 | 0.33 |
ENST00000471698.1 ENST00000189444.6 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr15_-_34629922 | 0.33 |
ENST00000559484.1 ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr6_+_24495067 | 0.33 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr2_-_165697920 | 0.32 |
ENST00000342193.4 ENST00000375458.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr7_+_29519662 | 0.32 |
ENST00000424025.2 ENST00000439711.2 ENST00000421775.2 |
CHN2 |
chimerin 2 |
| chr2_-_39347524 | 0.32 |
ENST00000395038.2 ENST00000402219.2 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr16_-_57513657 | 0.32 |
ENST00000566936.1 ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4 |
docking protein 4 |
| chr11_-_71159458 | 0.32 |
ENST00000355527.3 |
DHCR7 |
7-dehydrocholesterol reductase |
| chr2_-_220173685 | 0.32 |
ENST00000423636.2 ENST00000442029.1 ENST00000412847.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr2_-_32236002 | 0.32 |
ENST00000404530.1 |
MEMO1 |
mediator of cell motility 1 |
| chr2_-_165477971 | 0.31 |
ENST00000446413.2 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr7_+_73242490 | 0.31 |
ENST00000431918.1 |
CLDN4 |
claudin 4 |
| chr12_-_112819896 | 0.31 |
ENST00000377560.5 ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr3_+_38495333 | 0.31 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr19_-_51456198 | 0.31 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr14_-_104313824 | 0.31 |
ENST00000553739.1 ENST00000202556.9 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
| chr15_+_64752927 | 0.30 |
ENST00000416172.1 |
ZNF609 |
zinc finger protein 609 |
| chr18_-_45935663 | 0.30 |
ENST00000589194.1 ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
| chr2_-_27938593 | 0.30 |
ENST00000379677.2 |
AC074091.13 |
Uncharacterized protein |
| chr19_-_51456321 | 0.30 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr10_+_76871454 | 0.30 |
ENST00000372687.4 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr6_+_43543942 | 0.29 |
ENST00000372226.1 ENST00000443535.1 |
POLH |
polymerase (DNA directed), eta |
| chr6_+_43543864 | 0.29 |
ENST00000372236.4 ENST00000535400.1 |
POLH |
polymerase (DNA directed), eta |
| chr13_+_98628886 | 0.29 |
ENST00000490680.1 ENST00000539640.1 ENST00000403772.3 |
IPO5 |
importin 5 |
| chr22_-_31741757 | 0.28 |
ENST00000215919.3 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr17_+_77681075 | 0.28 |
ENST00000397549.2 |
CTD-2116F7.1 |
CTD-2116F7.1 |
| chr10_+_76871353 | 0.28 |
ENST00000542569.1 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr19_+_18284477 | 0.28 |
ENST00000407280.3 |
IFI30 |
interferon, gamma-inducible protein 30 |
| chr13_-_52027134 | 0.28 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr11_-_73309228 | 0.28 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr14_+_60716159 | 0.28 |
ENST00000325658.3 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_+_24272543 | 0.27 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr12_-_6798410 | 0.26 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr15_-_34630234 | 0.26 |
ENST00000558667.1 ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr7_+_65670186 | 0.26 |
ENST00000304842.5 ENST00000442120.1 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
| chr15_+_73344791 | 0.26 |
ENST00000261908.6 |
NEO1 |
neogenin 1 |
| chrX_+_51927919 | 0.26 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr1_+_37940153 | 0.26 |
ENST00000373087.6 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
| chr12_+_53773944 | 0.26 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr12_-_6798523 | 0.26 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr19_-_47249679 | 0.26 |
ENST00000263280.6 |
STRN4 |
striatin, calmodulin binding protein 4 |
| chr2_-_61697862 | 0.26 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr12_-_6798616 | 0.25 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr3_-_105587879 | 0.25 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr10_+_28822236 | 0.25 |
ENST00000347934.4 ENST00000354911.4 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr11_-_46142948 | 0.24 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr2_+_24272576 | 0.24 |
ENST00000380986.4 ENST00000452109.1 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr19_+_10531150 | 0.24 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr6_+_111580508 | 0.24 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chr1_+_203764742 | 0.23 |
ENST00000432282.1 ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A |
zinc finger CCCH-type containing 11A |
| chr11_+_119076745 | 0.22 |
ENST00000264033.4 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr19_-_38806560 | 0.22 |
ENST00000591755.1 ENST00000337679.8 ENST00000339413.6 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_-_77531858 | 0.22 |
ENST00000360355.2 |
RSF1 |
remodeling and spacing factor 1 |
| chr20_-_61493115 | 0.22 |
ENST00000335351.3 ENST00000217162.5 |
TCFL5 |
transcription factor-like 5 (basic helix-loop-helix) |
| chr3_-_188665428 | 0.22 |
ENST00000444488.1 |
TPRG1-AS1 |
TPRG1 antisense RNA 1 |
| chr2_-_172017393 | 0.22 |
ENST00000442919.2 |
TLK1 |
tousled-like kinase 1 |
| chr17_+_75277492 | 0.22 |
ENST00000427177.1 ENST00000591198.1 |
SEPT9 |
septin 9 |
| chr8_-_29120580 | 0.21 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr17_+_65821780 | 0.21 |
ENST00000321892.4 ENST00000335221.5 ENST00000306378.6 |
BPTF |
bromodomain PHD finger transcription factor |
| chr11_-_65325664 | 0.21 |
ENST00000301873.5 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr5_+_140254884 | 0.21 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr8_-_29208183 | 0.21 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr14_+_58765103 | 0.20 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr14_+_55738021 | 0.20 |
ENST00000313833.4 |
FBXO34 |
F-box protein 34 |
| chr2_+_232573208 | 0.20 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr7_-_100493482 | 0.19 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr18_+_3449821 | 0.19 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr22_+_29469100 | 0.19 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr2_-_176866978 | 0.19 |
ENST00000392540.2 ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715 |
KIAA1715 |
| chr3_+_111718173 | 0.19 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr17_+_73043301 | 0.19 |
ENST00000322444.6 |
KCTD2 |
potassium channel tetramerization domain containing 2 |
| chr19_-_14049184 | 0.18 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr2_+_28974603 | 0.18 |
ENST00000441461.1 ENST00000358506.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr20_-_62601218 | 0.18 |
ENST00000369888.1 |
ZNF512B |
zinc finger protein 512B |
| chr14_+_92980111 | 0.18 |
ENST00000216487.7 ENST00000557762.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr5_-_131132658 | 0.17 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr19_+_35759824 | 0.17 |
ENST00000343550.5 |
USF2 |
upstream transcription factor 2, c-fos interacting |
| chr3_-_149293990 | 0.17 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr11_+_117049445 | 0.17 |
ENST00000324225.4 ENST00000532960.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chrX_+_53078273 | 0.17 |
ENST00000332582.4 |
GPR173 |
G protein-coupled receptor 173 |
| chr5_+_110559784 | 0.17 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr2_-_25564750 | 0.17 |
ENST00000321117.5 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr5_+_110559603 | 0.17 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr14_-_105262055 | 0.17 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr10_+_8096769 | 0.16 |
ENST00000346208.3 |
GATA3 |
GATA binding protein 3 |
| chr7_+_100318423 | 0.16 |
ENST00000252723.2 |
EPO |
erythropoietin |
| chrX_+_16737718 | 0.16 |
ENST00000380155.3 |
SYAP1 |
synapse associated protein 1 |
| chr17_-_79869340 | 0.16 |
ENST00000538936.2 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr3_-_124774802 | 0.16 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chrX_-_53713657 | 0.15 |
ENST00000262854.6 ENST00000218328.8 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr10_+_28822417 | 0.15 |
ENST00000428935.1 ENST00000420266.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr6_-_34664612 | 0.15 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr21_+_46825032 | 0.15 |
ENST00000400337.2 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr11_+_117049854 | 0.15 |
ENST00000278951.7 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr3_-_10547333 | 0.15 |
ENST00000383800.4 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr1_+_22778337 | 0.14 |
ENST00000404138.1 ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40 |
zinc finger and BTB domain containing 40 |
| chr20_-_62710832 | 0.14 |
ENST00000395042.1 |
RGS19 |
regulator of G-protein signaling 19 |
| chr2_+_28974668 | 0.14 |
ENST00000296122.6 ENST00000395366.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_+_242254679 | 0.14 |
ENST00000428282.1 ENST00000360051.3 |
SEPT2 |
septin 2 |
| chr16_+_74411776 | 0.14 |
ENST00000429990.1 |
NPIPB15 |
nuclear pore complex interacting protein family, member B15 |
| chr14_+_106355918 | 0.14 |
ENST00000414005.1 |
AL122127.25 |
AL122127.25 |
| chr14_+_105886275 | 0.14 |
ENST00000405646.1 |
MTA1 |
metastasis associated 1 |
| chr8_+_81397876 | 0.14 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr13_+_25670268 | 0.13 |
ENST00000281589.3 |
PABPC3 |
poly(A) binding protein, cytoplasmic 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.1 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.3 | 1.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 1.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 1.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 2.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 2.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.9 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.1 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 3.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 1.6 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 0.9 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 0.9 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 1.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 1.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.7 | GO:1905064 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.2 | 0.5 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 1.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.6 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.6 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.7 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.6 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.3 | GO:0044828 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.5 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.4 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.1 | 0.7 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.1 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.2 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 2.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.0 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 0.5 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.1 | 0.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.2 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.7 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.3 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.6 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.8 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.0 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 2.1 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) |
| 0.0 | 0.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |