Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
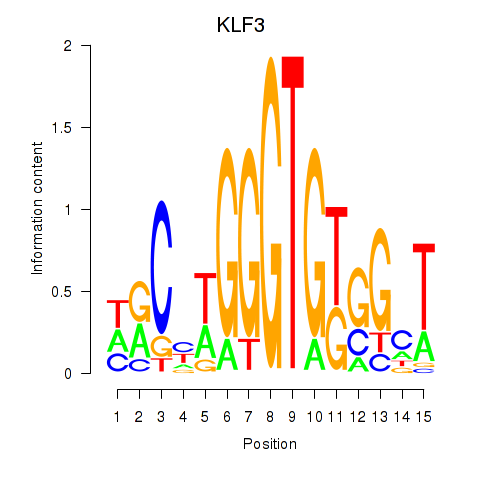
Results for KLF3
Z-value: 0.59
Transcription factors associated with KLF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF3
|
ENSG00000109787.8 | KLF3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF3 | hg19_v2_chr4_+_38665810_38665827 | -0.34 | 1.9e-01 | Click! |
Activity profile of KLF3 motif
Sorted Z-values of KLF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_152486108 | 1.96 |
ENST00000356661.5 |
MAGEA1 |
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
| chrX_+_52511925 | 1.12 |
ENST00000375588.1 |
XAGE1C |
X antigen family, member 1C |
| chrX_-_52260199 | 1.12 |
ENST00000375600.1 |
XAGE1A |
X antigen family, member 1A |
| chrX_-_52533139 | 1.11 |
ENST00000374959.3 |
XAGE1D |
X antigen family, member 1D |
| chrX_-_151938171 | 1.00 |
ENST00000393902.3 ENST00000417212.1 ENST00000370278.3 |
MAGEA3 |
melanoma antigen family A, 3 |
| chrX_-_52546033 | 0.98 |
ENST00000375567.3 |
XAGE1E |
X antigen family, member 1E |
| chr11_+_59480899 | 0.87 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr9_-_130637244 | 0.87 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr19_-_13213662 | 0.86 |
ENST00000264824.4 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
| chr11_-_414948 | 0.85 |
ENST00000530494.1 ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr11_-_61658853 | 0.77 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr19_-_40971667 | 0.74 |
ENST00000263368.4 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr19_-_40971643 | 0.68 |
ENST00000595483.1 |
BLVRB |
biliverdin reductase B (flavin reductase (NADPH)) |
| chr8_-_101963482 | 0.67 |
ENST00000419477.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr22_-_22874610 | 0.67 |
ENST00000302097.3 |
ZNF280A |
zinc finger protein 280A |
| chr15_-_58571445 | 0.66 |
ENST00000558231.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr1_+_151030234 | 0.64 |
ENST00000368921.3 |
MLLT11 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 |
| chr1_+_45477901 | 0.63 |
ENST00000434478.1 |
UROD |
uroporphyrinogen decarboxylase |
| chr11_-_61659006 | 0.62 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chrX_-_52260355 | 0.61 |
ENST00000375602.1 ENST00000399800.3 |
XAGE1A |
X antigen family, member 1A |
| chrX_+_52511761 | 0.59 |
ENST00000399795.3 ENST00000375589.1 |
XAGE1C |
X antigen family, member 1C |
| chr15_+_43985084 | 0.58 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr12_+_29302119 | 0.58 |
ENST00000536681.3 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr11_-_57194948 | 0.57 |
ENST00000533235.1 ENST00000526621.1 ENST00000352187.1 |
SLC43A3 |
solute carrier family 43, member 3 |
| chr4_-_144940477 | 0.54 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr15_+_43885252 | 0.54 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr14_-_21491305 | 0.54 |
ENST00000554531.1 |
NDRG2 |
NDRG family member 2 |
| chrX_+_151883090 | 0.51 |
ENST00000370293.2 ENST00000423993.1 ENST00000447530.1 ENST00000458057.1 ENST00000331220.2 ENST00000422085.1 ENST00000453150.1 ENST00000409560.1 |
MAGEA2B |
melanoma antigen family A, 2B |
| chr8_-_101962777 | 0.48 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_+_79989500 | 0.48 |
ENST00000306897.4 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chrX_-_52546189 | 0.46 |
ENST00000375570.1 ENST00000429372.2 |
XAGE1E |
X antigen family, member 1E |
| chrX_+_52238810 | 0.46 |
ENST00000437949.2 ENST00000375616.1 |
XAGE1B |
X antigen family, member 1B |
| chr15_-_60690932 | 0.44 |
ENST00000559818.1 |
ANXA2 |
annexin A2 |
| chr16_+_1578674 | 0.44 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr7_+_100466433 | 0.44 |
ENST00000429658.1 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr11_-_57158109 | 0.44 |
ENST00000525955.1 ENST00000533605.1 ENST00000311862.5 |
PRG2 |
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chr1_+_45477819 | 0.43 |
ENST00000246337.4 |
UROD |
uroporphyrinogen decarboxylase |
| chrX_+_153813407 | 0.43 |
ENST00000443287.2 ENST00000333128.3 |
CTAG1A |
cancer/testis antigen 1A |
| chrX_-_52533295 | 0.42 |
ENST00000375578.1 ENST00000396497.3 |
XAGE1D |
X antigen family, member 1D |
| chr10_+_45495898 | 0.42 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr12_+_48516357 | 0.41 |
ENST00000549022.1 ENST00000547587.1 ENST00000312352.7 |
PFKM |
phosphofructokinase, muscle |
| chr11_-_5248294 | 0.39 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr14_-_21492251 | 0.39 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_21492113 | 0.38 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr11_+_60048129 | 0.37 |
ENST00000355131.3 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr11_+_60048053 | 0.37 |
ENST00000337908.4 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr14_-_21491477 | 0.37 |
ENST00000298684.5 ENST00000557169.1 ENST00000553563.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_105635090 | 0.36 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr11_-_2906979 | 0.36 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chrX_-_71526999 | 0.36 |
ENST00000453707.2 ENST00000373619.3 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr2_+_37571845 | 0.35 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr22_+_42949925 | 0.35 |
ENST00000327678.5 ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2 |
serine hydrolase-like 2 |
| chrX_-_71526813 | 0.35 |
ENST00000246139.5 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr5_+_134240588 | 0.34 |
ENST00000254908.6 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr2_+_37571717 | 0.34 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr14_+_29234870 | 0.34 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr1_-_202130702 | 0.33 |
ENST00000309017.3 ENST00000477554.1 ENST00000492451.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr11_-_64512469 | 0.33 |
ENST00000377485.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_64512273 | 0.32 |
ENST00000377497.3 ENST00000377487.1 ENST00000430645.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr15_+_41523417 | 0.32 |
ENST00000560397.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr10_+_94451574 | 0.32 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr17_+_33914460 | 0.32 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr22_-_19842330 | 0.32 |
ENST00000416337.1 ENST00000328554.4 ENST00000403325.1 ENST00000453108.1 |
C22orf29 GNB1L |
chromosome 22 open reading frame 29 guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr19_+_19303572 | 0.32 |
ENST00000407360.3 ENST00000540981.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chrX_+_117480036 | 0.32 |
ENST00000371822.5 ENST00000254029.3 ENST00000371825.3 |
WDR44 |
WD repeat domain 44 |
| chr12_-_12503156 | 0.32 |
ENST00000543314.1 ENST00000396349.3 |
MANSC1 |
MANSC domain containing 1 |
| chr15_+_43985725 | 0.31 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr6_+_150070857 | 0.31 |
ENST00000544496.1 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr22_+_29702996 | 0.31 |
ENST00000406549.3 ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr17_-_3704500 | 0.31 |
ENST00000263087.4 |
ITGAE |
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
| chr4_-_90757364 | 0.31 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_+_19303008 | 0.30 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr19_-_3606590 | 0.30 |
ENST00000411851.3 |
TBXA2R |
thromboxane A2 receptor |
| chr11_+_844067 | 0.29 |
ENST00000397406.1 ENST00000409543.2 ENST00000525201.1 |
TSPAN4 |
tetraspanin 4 |
| chr22_+_29702572 | 0.29 |
ENST00000407647.2 ENST00000416823.1 ENST00000428622.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr7_+_116165754 | 0.29 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr6_+_150070831 | 0.29 |
ENST00000367380.5 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr17_-_73505961 | 0.29 |
ENST00000433559.2 |
CASKIN2 |
CASK interacting protein 2 |
| chr19_-_10227503 | 0.29 |
ENST00000593054.1 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
| chr12_+_122326662 | 0.28 |
ENST00000261817.2 ENST00000538613.1 ENST00000542602.1 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr22_-_26961328 | 0.27 |
ENST00000398110.2 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
| chr1_+_180165672 | 0.27 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_+_25870070 | 0.27 |
ENST00000374338.4 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
| chr2_-_158732340 | 0.27 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr11_+_8932715 | 0.26 |
ENST00000529876.1 ENST00000525005.1 ENST00000524577.1 ENST00000534506.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr17_+_33914276 | 0.26 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr1_+_207494853 | 0.26 |
ENST00000367064.3 ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr11_-_14913190 | 0.26 |
ENST00000532378.1 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr10_+_60028818 | 0.25 |
ENST00000333926.5 |
CISD1 |
CDGSH iron sulfur domain 1 |
| chr15_-_83876758 | 0.25 |
ENST00000299633.4 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
| chr17_+_38599693 | 0.25 |
ENST00000542955.1 ENST00000269593.4 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
| chr7_-_73153122 | 0.25 |
ENST00000458339.1 |
ABHD11 |
abhydrolase domain containing 11 |
| chr2_+_172378757 | 0.25 |
ENST00000409484.1 ENST00000321348.4 ENST00000375252.3 |
CYBRD1 |
cytochrome b reductase 1 |
| chr17_-_79980734 | 0.25 |
ENST00000584600.1 ENST00000584347.1 ENST00000580435.1 ENST00000306704.6 ENST00000392359.3 |
STRA13 |
stimulated by retinoic acid 13 |
| chr11_+_112097069 | 0.25 |
ENST00000280362.3 ENST00000525803.1 |
PTS |
6-pyruvoyltetrahydropterin synthase |
| chr6_-_106773291 | 0.25 |
ENST00000343245.3 |
ATG5 |
autophagy related 5 |
| chr22_+_39760130 | 0.24 |
ENST00000381535.4 |
SYNGR1 |
synaptogyrin 1 |
| chr7_-_73153178 | 0.24 |
ENST00000437775.2 ENST00000222800.3 |
ABHD11 |
abhydrolase domain containing 11 |
| chr8_-_101964231 | 0.24 |
ENST00000521309.1 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_+_122326630 | 0.23 |
ENST00000541212.1 ENST00000340175.5 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr11_+_844406 | 0.23 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr19_-_55919087 | 0.23 |
ENST00000587845.1 ENST00000589978.1 ENST00000264552.9 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
| chrX_-_71933888 | 0.23 |
ENST00000373542.4 ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
| chr6_-_106773610 | 0.23 |
ENST00000369076.3 ENST00000369070.1 |
ATG5 |
autophagy related 5 |
| chr8_-_145642267 | 0.23 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr22_+_31523734 | 0.23 |
ENST00000402238.1 ENST00000404453.1 ENST00000401755.1 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr15_+_43886057 | 0.23 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr8_-_144699628 | 0.22 |
ENST00000529048.1 ENST00000529064.1 |
TSTA3 |
tissue specific transplantation antigen P35B |
| chr16_-_67514982 | 0.22 |
ENST00000565835.1 ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chrX_+_129473916 | 0.22 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr4_-_681114 | 0.22 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr15_+_63334831 | 0.22 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr8_+_59465728 | 0.21 |
ENST00000260130.4 ENST00000422546.2 ENST00000447182.2 ENST00000413219.2 ENST00000424270.2 ENST00000523483.1 ENST00000520168.1 |
SDCBP |
syndecan binding protein (syntenin) |
| chr17_+_27181956 | 0.21 |
ENST00000254928.5 ENST00000580917.2 |
ERAL1 |
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr3_-_171178157 | 0.21 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
| chr7_-_148580563 | 0.21 |
ENST00000476773.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr11_-_64511575 | 0.21 |
ENST00000431822.1 ENST00000377486.3 ENST00000394432.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr7_-_73153161 | 0.21 |
ENST00000395147.4 |
ABHD11 |
abhydrolase domain containing 11 |
| chr12_-_113772835 | 0.21 |
ENST00000552014.1 ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr19_+_41107249 | 0.21 |
ENST00000396819.3 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr17_-_19290483 | 0.21 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr11_+_8932654 | 0.21 |
ENST00000299576.5 ENST00000309377.4 ENST00000309357.4 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr18_+_9334755 | 0.21 |
ENST00000262120.5 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
| chr11_-_64512803 | 0.20 |
ENST00000377489.1 ENST00000354024.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_114466477 | 0.20 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr1_-_6445809 | 0.20 |
ENST00000377855.2 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr12_-_15114492 | 0.20 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_21927736 | 0.20 |
ENST00000240662.2 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr7_-_76255444 | 0.20 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr3_+_54156570 | 0.20 |
ENST00000415676.2 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr7_-_102184083 | 0.20 |
ENST00000379357.5 |
POLR2J3 |
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr11_+_8932828 | 0.20 |
ENST00000530281.1 ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chrX_-_153599578 | 0.19 |
ENST00000360319.4 ENST00000344736.4 |
FLNA |
filamin A, alpha |
| chr3_+_54156664 | 0.19 |
ENST00000474759.1 ENST00000288197.5 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr1_+_95699704 | 0.19 |
ENST00000370202.4 |
RWDD3 |
RWD domain containing 3 |
| chr20_-_590944 | 0.19 |
ENST00000246080.3 |
TCF15 |
transcription factor 15 (basic helix-loop-helix) |
| chr19_-_3025614 | 0.19 |
ENST00000447365.2 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr4_-_106395135 | 0.19 |
ENST00000310267.7 |
PPA2 |
pyrophosphatase (inorganic) 2 |
| chr15_+_67430339 | 0.19 |
ENST00000439724.3 |
SMAD3 |
SMAD family member 3 |
| chr1_+_210001309 | 0.19 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr4_+_26585538 | 0.18 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr19_+_50919056 | 0.18 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr11_-_111783919 | 0.18 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr6_-_106773491 | 0.18 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr11_-_62313090 | 0.18 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr3_+_49449636 | 0.18 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr12_+_116997186 | 0.18 |
ENST00000306985.4 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
| chr3_+_11196206 | 0.18 |
ENST00000431010.2 |
HRH1 |
histamine receptor H1 |
| chr11_-_535515 | 0.18 |
ENST00000311189.7 ENST00000451590.1 ENST00000417302.1 |
HRAS |
Harvey rat sarcoma viral oncogene homolog |
| chr20_+_47662805 | 0.18 |
ENST00000262982.2 ENST00000542325.1 |
CSE1L |
CSE1 chromosome segregation 1-like (yeast) |
| chr17_-_58096336 | 0.17 |
ENST00000587125.1 ENST00000407042.3 |
TBC1D3P1-DHX40P1 |
TBC1D3P1-DHX40P1 readthrough transcribed pseudogene |
| chr2_+_231921574 | 0.17 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr2_+_48757278 | 0.17 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr8_-_101964265 | 0.17 |
ENST00000395958.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_+_20081515 | 0.17 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr13_-_99630233 | 0.16 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr16_+_66400533 | 0.16 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr17_-_73149921 | 0.16 |
ENST00000481647.1 ENST00000470924.1 |
HN1 |
hematological and neurological expressed 1 |
| chr5_+_151151471 | 0.16 |
ENST00000394123.3 ENST00000543466.1 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_+_85061367 | 0.16 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr19_-_53193731 | 0.16 |
ENST00000598536.1 ENST00000594682.2 ENST00000601257.1 |
ZNF83 |
zinc finger protein 83 |
| chr18_+_61554932 | 0.16 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr7_-_752577 | 0.16 |
ENST00000544935.1 ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_+_75970672 | 0.15 |
ENST00000435356.1 |
AC105020.1 |
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr12_-_118797475 | 0.15 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr19_-_4455290 | 0.15 |
ENST00000394765.3 ENST00000592515.1 |
UBXN6 |
UBX domain protein 6 |
| chr1_-_205744205 | 0.15 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr15_-_28344439 | 0.15 |
ENST00000431101.1 ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2 |
oculocutaneous albinism II |
| chr15_-_85259360 | 0.15 |
ENST00000559729.1 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
| chr1_-_38273840 | 0.15 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr12_+_93861282 | 0.15 |
ENST00000552217.1 ENST00000393128.4 ENST00000547098.1 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr8_-_13134045 | 0.15 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr11_+_34073269 | 0.14 |
ENST00000389645.3 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr6_-_28891709 | 0.14 |
ENST00000377194.3 ENST00000377199.3 |
TRIM27 |
tripartite motif containing 27 |
| chr8_+_145582633 | 0.14 |
ENST00000540505.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr1_+_95699740 | 0.14 |
ENST00000429514.2 ENST00000263893.6 |
RWDD3 |
RWD domain containing 3 |
| chr2_+_133174147 | 0.14 |
ENST00000329321.3 |
GPR39 |
G protein-coupled receptor 39 |
| chr19_+_46850320 | 0.14 |
ENST00000391919.1 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr22_+_32340481 | 0.14 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chrX_-_100604184 | 0.14 |
ENST00000372902.3 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chrX_-_47518527 | 0.14 |
ENST00000333119.3 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
| chr12_+_21525818 | 0.14 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr2_-_85637459 | 0.14 |
ENST00000409921.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr11_-_72432950 | 0.14 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr19_-_44174330 | 0.14 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr7_-_102283238 | 0.13 |
ENST00000340457.8 |
UPK3BL |
uroplakin 3B-like |
| chr11_-_65626753 | 0.13 |
ENST00000526975.1 ENST00000531413.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chrX_+_107069063 | 0.13 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr6_+_34725181 | 0.13 |
ENST00000244520.5 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr8_-_145013711 | 0.13 |
ENST00000345136.3 |
PLEC |
plectin |
| chr4_-_168155417 | 0.13 |
ENST00000511269.1 ENST00000506697.1 ENST00000512042.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_110453514 | 0.13 |
ENST00000369802.3 ENST00000420111.2 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr11_-_59578202 | 0.13 |
ENST00000300151.4 |
MRPL16 |
mitochondrial ribosomal protein L16 |
| chr1_+_213224572 | 0.13 |
ENST00000543470.1 ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr1_+_24882560 | 0.13 |
ENST00000374392.2 |
NCMAP |
noncompact myelin associated protein |
| chr12_+_44152740 | 0.13 |
ENST00000440781.2 ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4 |
interleukin-1 receptor-associated kinase 4 |
| chr19_+_41313017 | 0.12 |
ENST00000595621.1 ENST00000595051.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr9_-_34710066 | 0.12 |
ENST00000378792.1 ENST00000259607.2 |
CCL21 |
chemokine (C-C motif) ligand 21 |
| chr12_+_93861264 | 0.12 |
ENST00000549982.1 ENST00000361630.2 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr22_+_32340447 | 0.12 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.0 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 0.7 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.6 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.3 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.1 | 1.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 1.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.3 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 0.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.4 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 1.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.2 | GO:0042351 | GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 1.0 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:2000561 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.1 | 0.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.2 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 2.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.4 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 0.4 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:1902228 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.2 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.5 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.2 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.2 | GO:0016479 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0048852 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.4 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.4 | 1.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.2 | 0.7 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.3 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 1.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.9 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 2.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |