Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
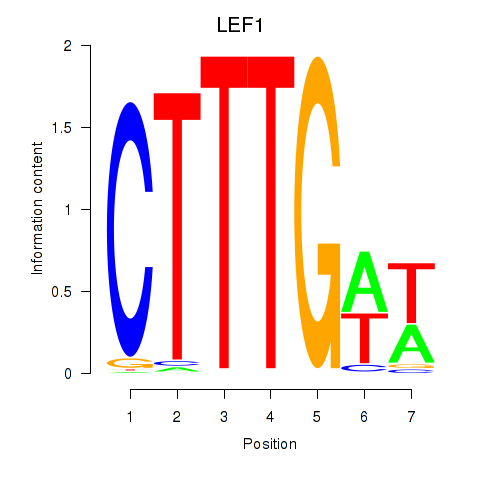
Results for LEF1
Z-value: 1.03
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | LEF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109087872_109087881, hg19_v2_chr4_-_109088940_109089037 | -0.34 | 1.9e-01 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_54074033 | 4.00 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr15_+_96869165 | 2.61 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_149375783 | 2.26 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr7_+_16793160 | 2.06 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr12_-_6233828 | 1.77 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr2_-_216257849 | 1.75 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr11_+_69455855 | 1.59 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr5_+_66124590 | 1.52 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_15518923 | 1.52 |
ENST00000348343.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr5_+_34757309 | 1.51 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr1_+_100111479 | 1.46 |
ENST00000263174.4 |
PALMD |
palmdelphin |
| chr13_-_72440901 | 1.45 |
ENST00000359684.2 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr11_+_114128522 | 1.41 |
ENST00000535401.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr1_+_93913713 | 1.41 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chr13_-_110959478 | 1.36 |
ENST00000543140.1 ENST00000375820.4 |
COL4A1 |
collagen, type IV, alpha 1 |
| chr14_-_21270995 | 1.32 |
ENST00000555698.1 ENST00000397970.4 ENST00000340900.3 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr1_+_100111580 | 1.32 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chr3_-_134093395 | 1.32 |
ENST00000249883.5 |
AMOTL2 |
angiomotin like 2 |
| chr4_-_186877502 | 1.31 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_-_1748214 | 1.31 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr3_-_169381183 | 1.20 |
ENST00000494292.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr10_-_95241951 | 1.19 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr10_-_95242044 | 1.18 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr8_-_17579726 | 1.18 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr3_-_99569821 | 1.18 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr13_-_72441315 | 1.14 |
ENST00000305425.4 ENST00000313174.7 ENST00000354591.4 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr6_+_143929307 | 1.10 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr3_+_173302222 | 1.08 |
ENST00000361589.4 |
NLGN1 |
neuroligin 1 |
| chr5_-_16936340 | 1.08 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr3_-_134093275 | 1.06 |
ENST00000513145.1 ENST00000422605.2 |
AMOTL2 |
angiomotin like 2 |
| chr20_-_23066953 | 1.06 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr9_+_504674 | 1.05 |
ENST00000382297.2 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr18_+_3449821 | 1.04 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr7_-_27213893 | 1.01 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr15_+_63335899 | 0.96 |
ENST00000561266.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr7_-_27224842 | 0.94 |
ENST00000517402.1 |
HOXA11 |
homeobox A11 |
| chr18_-_25616519 | 0.94 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_-_157892498 | 0.89 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr7_-_27224795 | 0.88 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr12_+_6309963 | 0.87 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr7_-_27219849 | 0.86 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr14_+_85996471 | 0.78 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr15_+_96873921 | 0.77 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_77924627 | 0.75 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr2_-_165424973 | 0.75 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr10_+_114710211 | 0.72 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr22_-_36357671 | 0.72 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr20_+_11898507 | 0.70 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr1_-_12677714 | 0.70 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr17_-_46623441 | 0.70 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr18_+_55888767 | 0.69 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_-_105444694 | 0.67 |
ENST00000333244.5 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr4_-_139163491 | 0.66 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr13_-_45150392 | 0.65 |
ENST00000501704.2 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr6_-_134639180 | 0.65 |
ENST00000367858.5 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chr5_-_146833485 | 0.65 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr18_-_52989217 | 0.63 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chrX_-_10588459 | 0.63 |
ENST00000380782.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr1_-_225840747 | 0.62 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr5_+_131593364 | 0.61 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr14_+_85996507 | 0.61 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr2_+_181845843 | 0.61 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr10_-_30024716 | 0.60 |
ENST00000375398.2 ENST00000375400.3 |
SVIL |
supervillin |
| chr6_-_112575912 | 0.58 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr2_-_175629135 | 0.58 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_82266053 | 0.58 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr9_+_124088860 | 0.57 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr17_-_3571934 | 0.57 |
ENST00000225525.3 |
TAX1BP3 |
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr10_+_123923205 | 0.57 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_57673430 | 0.57 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chrX_-_10588595 | 0.56 |
ENST00000423614.1 ENST00000317552.4 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr10_+_71561630 | 0.56 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr11_-_6677018 | 0.55 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr11_+_101983176 | 0.54 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr8_+_39770803 | 0.54 |
ENST00000518237.1 |
IDO1 |
indoleamine 2,3-dioxygenase 1 |
| chr7_+_107224364 | 0.54 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr10_-_69597915 | 0.54 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr18_-_53089723 | 0.53 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr10_+_114709999 | 0.53 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_-_16900217 | 0.53 |
ENST00000441778.2 |
LDB2 |
LIM domain binding 2 |
| chr4_-_16900410 | 0.52 |
ENST00000304523.5 |
LDB2 |
LIM domain binding 2 |
| chr4_-_16900242 | 0.51 |
ENST00000502640.1 ENST00000506732.1 |
LDB2 |
LIM domain binding 2 |
| chr4_-_16900184 | 0.51 |
ENST00000515064.1 |
LDB2 |
LIM domain binding 2 |
| chr20_-_56284816 | 0.50 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr15_+_50474385 | 0.50 |
ENST00000267842.5 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chr13_-_67802549 | 0.50 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr13_-_99630233 | 0.50 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr19_-_3028354 | 0.49 |
ENST00000586422.1 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr13_+_32605437 | 0.49 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr4_+_20255123 | 0.48 |
ENST00000504154.1 ENST00000273739.5 |
SLIT2 |
slit homolog 2 (Drosophila) |
| chr12_+_79258547 | 0.48 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr1_+_196621002 | 0.46 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr19_-_6690723 | 0.46 |
ENST00000601008.1 |
C3 |
complement component 3 |
| chr2_+_85360499 | 0.46 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr7_+_116312411 | 0.46 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr10_+_71561649 | 0.46 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr1_+_66797687 | 0.45 |
ENST00000371045.5 ENST00000531025.1 ENST00000526197.1 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr6_-_112575838 | 0.45 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr10_+_71561704 | 0.44 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr9_+_103235365 | 0.44 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chrX_+_54835493 | 0.43 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr10_-_69597810 | 0.43 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr17_-_46682321 | 0.43 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr11_-_64013663 | 0.43 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr15_-_23932437 | 0.42 |
ENST00000331837.4 |
NDN |
necdin, melanoma antigen (MAGE) family member |
| chr2_-_27592867 | 0.42 |
ENST00000451130.2 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr4_-_85887503 | 0.41 |
ENST00000509172.1 ENST00000322366.6 ENST00000295888.4 ENST00000502713.1 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
| chr7_+_73868439 | 0.41 |
ENST00000424337.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr1_+_84630053 | 0.40 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_175629164 | 0.40 |
ENST00000409323.1 ENST00000261007.5 ENST00000348749.5 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr11_+_57529234 | 0.40 |
ENST00000360682.6 ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
| chr4_+_146403912 | 0.39 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr2_-_27593306 | 0.38 |
ENST00000347454.4 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr3_-_58563094 | 0.38 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr16_+_68678892 | 0.36 |
ENST00000429102.2 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr2_-_27593180 | 0.36 |
ENST00000493344.2 ENST00000445933.2 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr1_+_155036204 | 0.36 |
ENST00000368409.3 ENST00000359751.4 ENST00000427683.2 ENST00000556931.1 ENST00000505139.1 |
EFNA4 EFNA3 EFNA3 |
ephrin-A4 ephrin-A3 Ephrin-A3; Uncharacterized protein; cDNA FLJ57652, highly similar to Ephrin-A3 |
| chrX_+_135229600 | 0.35 |
ENST00000370690.3 |
FHL1 |
four and a half LIM domains 1 |
| chr5_-_176900610 | 0.35 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chrX_-_10645773 | 0.34 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr11_-_75017734 | 0.34 |
ENST00000532525.1 |
ARRB1 |
arrestin, beta 1 |
| chr4_-_111119804 | 0.34 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr2_-_26205340 | 0.34 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr1_-_94079648 | 0.33 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chrX_+_37545012 | 0.33 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr11_+_65408273 | 0.33 |
ENST00000394227.3 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr2_+_234621551 | 0.32 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr9_-_94186131 | 0.31 |
ENST00000297689.3 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
| chr16_+_68678739 | 0.31 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr1_-_95392635 | 0.31 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chr1_-_211752073 | 0.31 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr9_+_36036430 | 0.30 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr10_+_114710425 | 0.30 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr18_+_29027696 | 0.30 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr1_+_6845384 | 0.30 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chrX_+_102631844 | 0.30 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr1_+_23037323 | 0.29 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr11_+_65339820 | 0.29 |
ENST00000316409.2 ENST00000449319.2 ENST00000530349.1 |
FAM89B |
family with sequence similarity 89, member B |
| chr7_-_27169801 | 0.29 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr5_+_137514687 | 0.29 |
ENST00000394894.3 |
KIF20A |
kinesin family member 20A |
| chr2_+_198669365 | 0.29 |
ENST00000428675.1 |
PLCL1 |
phospholipase C-like 1 |
| chr19_-_3029011 | 0.28 |
ENST00000590536.1 ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr9_-_23825956 | 0.28 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_52813932 | 0.28 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr5_+_137514834 | 0.28 |
ENST00000508792.1 ENST00000504621.1 |
KIF20A |
kinesin family member 20A |
| chr1_+_196621156 | 0.28 |
ENST00000359637.2 |
CFH |
complement factor H |
| chrX_+_51927919 | 0.27 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr7_+_73868220 | 0.27 |
ENST00000455841.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr7_-_140178775 | 0.26 |
ENST00000474576.1 ENST00000473444.1 ENST00000471104.1 |
MKRN1 |
makorin ring finger protein 1 |
| chr21_+_39628655 | 0.26 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr10_+_71562180 | 0.26 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr5_+_92919043 | 0.26 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_32345305 | 0.26 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chrX_+_135229559 | 0.26 |
ENST00000394155.2 |
FHL1 |
four and a half LIM domains 1 |
| chr4_+_30721968 | 0.25 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr11_-_64013288 | 0.25 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_-_33686743 | 0.25 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr14_+_61201445 | 0.25 |
ENST00000261245.4 ENST00000539616.2 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
| chrX_-_39956656 | 0.25 |
ENST00000397354.3 ENST00000378444.4 |
BCOR |
BCL6 corepressor |
| chrX_+_86772707 | 0.24 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chr15_+_43985725 | 0.24 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr12_+_9067123 | 0.24 |
ENST00000543824.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr7_-_41742697 | 0.24 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr1_+_100818156 | 0.23 |
ENST00000336454.3 |
CDC14A |
cell division cycle 14A |
| chr20_+_36149602 | 0.23 |
ENST00000062104.2 ENST00000346199.2 |
NNAT |
neuronatin |
| chr12_+_117013656 | 0.23 |
ENST00000556529.1 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
| chr21_+_39628852 | 0.23 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_-_106641728 | 0.23 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr6_-_32145861 | 0.23 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chrX_+_86772787 | 0.23 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr15_-_48470558 | 0.23 |
ENST00000324324.7 |
MYEF2 |
myelin expression factor 2 |
| chr6_+_32146131 | 0.22 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr18_-_52989525 | 0.22 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr17_-_62502022 | 0.22 |
ENST00000578804.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr4_+_113152881 | 0.22 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr3_-_171528227 | 0.22 |
ENST00000356327.5 ENST00000342215.6 ENST00000340989.4 ENST00000351298.4 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
| chr1_-_109935819 | 0.22 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr6_-_112575758 | 0.21 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr15_+_40731920 | 0.21 |
ENST00000561234.1 |
BAHD1 |
bromo adjacent homology domain containing 1 |
| chr11_+_120255997 | 0.21 |
ENST00000532993.1 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_68150744 | 0.21 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chrX_-_119694538 | 0.21 |
ENST00000371322.5 |
CUL4B |
cullin 4B |
| chr12_+_81110684 | 0.20 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr4_+_113152978 | 0.20 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr7_-_102985035 | 0.20 |
ENST00000426036.2 ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2 |
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr16_+_89989687 | 0.20 |
ENST00000315491.7 ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3 |
Tubulin beta-3 chain |
| chr12_-_108154925 | 0.20 |
ENST00000228437.5 |
PRDM4 |
PR domain containing 4 |
| chr12_+_53491220 | 0.20 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr12_-_54691668 | 0.20 |
ENST00000553198.1 |
NFE2 |
nuclear factor, erythroid 2 |
| chr10_+_97803151 | 0.20 |
ENST00000403870.3 ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ |
cyclin J |
| chr19_-_18717627 | 0.20 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr3_+_6902794 | 0.19 |
ENST00000357716.4 ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7 |
glutamate receptor, metabotropic 7 |
| chr12_-_95611149 | 0.19 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr2_-_161056762 | 0.19 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr1_-_244006528 | 0.19 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr17_-_56065484 | 0.19 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr2_-_100987007 | 0.19 |
ENST00000595083.1 |
AC012493.2 |
Uncharacterized protein |
| chr9_+_71820057 | 0.19 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr7_-_47621736 | 0.19 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr16_-_30798492 | 0.19 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.8 | 3.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 1.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.4 | 2.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 2.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 1.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.8 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.3 | 3.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 2.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.2 | 0.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.5 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 1.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.4 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.8 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 0.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 1.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.6 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 1.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:0060278 | negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 1.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.1 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 1.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 2.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 1.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.2 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.4 | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.6 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.7 | GO:0042325 | regulation of phosphorylation(GO:0042325) |
| 0.0 | 0.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.9 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 1.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 1.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 1.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 1.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 1.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 2.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 4.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 4.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 9.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 2.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.4 | 1.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 1.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 0.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 1.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 3.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 1.8 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 1.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |