Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
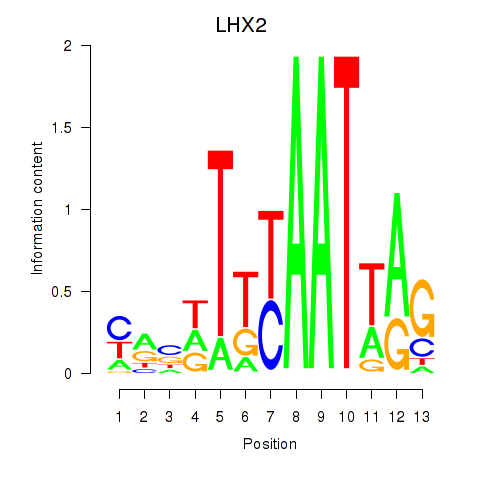
Results for LHX2
Z-value: 0.39
Transcription factors associated with LHX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX2
|
ENSG00000106689.6 | LHX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX2 | hg19_v2_chr9_+_126777676_126777700, hg19_v2_chr9_+_126773880_126773895 | 0.53 | 3.6e-02 | Click! |
Activity profile of LHX2 motif
Sorted Z-values of LHX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 1.14 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr13_-_99959641 | 1.03 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr12_+_6309517 | 0.69 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr11_-_104972158 | 0.67 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr14_-_106926724 | 0.61 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr2_-_89157161 | 0.60 |
ENST00000390237.2 |
IGKC |
immunoglobulin kappa constant |
| chr1_+_160709055 | 0.58 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr12_+_6554021 | 0.52 |
ENST00000266557.3 |
CD27 |
CD27 molecule |
| chr1_+_160709029 | 0.51 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chr13_-_99910673 | 0.39 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr4_-_90756769 | 0.38 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_-_183560011 | 0.34 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr2_+_234627424 | 0.34 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr4_-_153332886 | 0.29 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_-_105416039 | 0.27 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr2_-_231084617 | 0.26 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr6_+_30848557 | 0.25 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_68961905 | 0.24 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr18_+_34124507 | 0.24 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr2_+_68961934 | 0.24 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr7_-_5998714 | 0.23 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr11_-_104827425 | 0.23 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_125524785 | 0.22 |
ENST00000392482.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr13_+_73632897 | 0.21 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr2_-_231084659 | 0.20 |
ENST00000258381.6 ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110 |
SP110 nuclear body protein |
| chr15_-_72563585 | 0.19 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr10_-_14372870 | 0.18 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr16_+_31271274 | 0.18 |
ENST00000287497.8 ENST00000544665.3 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr4_-_120243545 | 0.17 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr2_-_10587897 | 0.17 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr12_-_118797475 | 0.15 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr11_+_64001962 | 0.14 |
ENST00000309422.2 |
VEGFB |
vascular endothelial growth factor B |
| chr18_+_3247779 | 0.14 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr21_+_34619079 | 0.13 |
ENST00000433395.2 |
AP000295.9 |
AP000295.9 |
| chr9_+_116207007 | 0.13 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr4_+_69313145 | 0.12 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr14_+_69865401 | 0.12 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr8_-_109260897 | 0.11 |
ENST00000521297.1 ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E |
eukaryotic translation initiation factor 3, subunit E |
| chr10_+_96443204 | 0.11 |
ENST00000339022.5 |
CYP2C18 |
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr7_-_54826920 | 0.11 |
ENST00000395535.3 ENST00000352861.4 |
SEC61G |
Sec61 gamma subunit |
| chr12_-_11548496 | 0.11 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr5_+_179921344 | 0.10 |
ENST00000261951.4 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr17_-_38859996 | 0.10 |
ENST00000264651.2 |
KRT24 |
keratin 24 |
| chr12_+_16064106 | 0.10 |
ENST00000428559.2 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr1_-_147142557 | 0.10 |
ENST00000369238.6 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
| chr14_-_51027838 | 0.08 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_-_103848405 | 0.08 |
ENST00000447452.2 ENST00000545943.1 ENST00000297431.4 |
ORC5 |
origin recognition complex, subunit 5 |
| chr3_+_108541545 | 0.08 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr7_-_54826869 | 0.07 |
ENST00000450622.1 |
SEC61G |
Sec61 gamma subunit |
| chr2_+_90139056 | 0.07 |
ENST00000492446.1 |
IGKV1D-16 |
immunoglobulin kappa variable 1D-16 |
| chr11_+_31391381 | 0.07 |
ENST00000465995.1 ENST00000536040.1 |
DNAJC24 |
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr11_+_27076764 | 0.07 |
ENST00000525090.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_64173583 | 0.07 |
ENST00000261234.6 |
TMEM5 |
transmembrane protein 5 |
| chr9_-_100854838 | 0.07 |
ENST00000538344.1 |
TRIM14 |
tripartite motif containing 14 |
| chr7_-_122840015 | 0.06 |
ENST00000194130.2 |
SLC13A1 |
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr2_+_210517895 | 0.06 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr17_-_39222131 | 0.06 |
ENST00000394015.2 |
KRTAP2-4 |
keratin associated protein 2-4 |
| chr3_+_108541608 | 0.06 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr13_+_33160553 | 0.06 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_-_107211459 | 0.05 |
ENST00000390636.2 |
IGHV3-73 |
immunoglobulin heavy variable 3-73 |
| chr9_+_108320392 | 0.05 |
ENST00000602661.1 ENST00000223528.2 ENST00000448551.2 ENST00000540160.1 |
FKTN |
fukutin |
| chr2_+_145780767 | 0.05 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr15_+_58430567 | 0.05 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr4_+_88532028 | 0.05 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr15_+_58430368 | 0.05 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr18_+_66465302 | 0.05 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr21_-_35014027 | 0.04 |
ENST00000399442.1 ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr2_+_45168875 | 0.04 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr6_-_26027480 | 0.04 |
ENST00000377364.3 |
HIST1H4B |
histone cluster 1, H4b |
| chr16_+_24266874 | 0.04 |
ENST00000005284.3 |
CACNG3 |
calcium channel, voltage-dependent, gamma subunit 3 |
| chr1_-_89591749 | 0.04 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chrX_+_99839799 | 0.04 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr8_+_76452097 | 0.03 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr9_+_105757590 | 0.03 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr19_+_22235310 | 0.03 |
ENST00000600162.1 |
ZNF257 |
zinc finger protein 257 |
| chr6_+_26199737 | 0.03 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr19_+_42212526 | 0.02 |
ENST00000598976.1 ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA CEACAM5 |
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr2_+_145780739 | 0.02 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr12_-_25348007 | 0.02 |
ENST00000354189.5 ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1 |
cancer susceptibility candidate 1 |
| chr11_+_24518723 | 0.02 |
ENST00000336930.6 ENST00000529015.1 ENST00000533227.1 |
LUZP2 |
leucine zipper protein 2 |
| chr12_-_10955226 | 0.02 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr3_-_131221790 | 0.02 |
ENST00000512877.1 ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3 |
mitochondrial ribosomal protein L3 |
| chr4_+_128554081 | 0.02 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr3_+_119187785 | 0.02 |
ENST00000295588.4 ENST00000476573.1 |
POGLUT1 |
protein O-glucosyltransferase 1 |
| chr13_-_19755975 | 0.02 |
ENST00000400113.3 |
TUBA3C |
tubulin, alpha 3c |
| chr4_+_70796784 | 0.02 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr7_-_14880892 | 0.02 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr2_-_208989225 | 0.02 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr18_-_19994830 | 0.01 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr14_+_22694606 | 0.01 |
ENST00000390463.3 |
TRAV36DV7 |
T cell receptor alpha variable 36/delta variable 7 |
| chr10_-_90712520 | 0.01 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr7_+_156433573 | 0.01 |
ENST00000311822.8 |
RNF32 |
ring finger protein 32 |
| chr2_+_173686303 | 0.01 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_56078695 | 0.00 |
ENST00000416613.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chrX_-_138724677 | 0.00 |
ENST00000370573.4 ENST00000338585.6 ENST00000370576.4 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr17_+_17082842 | 0.00 |
ENST00000579361.1 |
MPRIP |
myosin phosphatase Rho interacting protein |
| chr12_+_122688090 | 0.00 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr9_+_21440440 | 0.00 |
ENST00000276927.1 |
IFNA1 |
interferon, alpha 1 |
| chr12_+_94071129 | 0.00 |
ENST00000552983.1 ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD |
CASP2 and RIPK1 domain containing adaptor with death domain |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.0 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:2000468 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.4 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.1 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0021798 | neuroblast differentiation(GO:0014016) forebrain dorsal/ventral pattern formation(GO:0021798) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0045730 | respiratory burst(GO:0045730) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |