Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
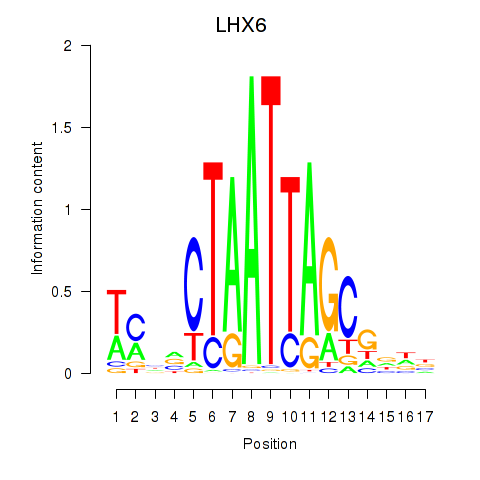
Results for LHX6
Z-value: 1.17
Transcription factors associated with LHX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX6
|
ENSG00000106852.11 | LHX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX6 | hg19_v2_chr9_-_124990680_124990719 | -0.60 | 1.3e-02 | Click! |
Activity profile of LHX6 motif
Sorted Z-values of LHX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_52535712 | 4.30 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr20_+_19867150 | 3.77 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr11_+_5710919 | 3.69 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr6_-_167040731 | 3.31 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr16_+_55512742 | 2.85 |
ENST00000568715.1 ENST00000219070.4 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr11_-_102668879 | 2.55 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr3_-_149095652 | 2.53 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr3_-_99569821 | 2.48 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr12_-_91546926 | 1.87 |
ENST00000550758.1 |
DCN |
decorin |
| chr10_+_13141441 | 1.66 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr21_+_35014783 | 1.51 |
ENST00000381291.4 ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr10_+_13141585 | 1.49 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr5_-_54830871 | 1.49 |
ENST00000307259.8 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr1_+_81771806 | 1.43 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr12_-_102455902 | 1.42 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr5_-_54830784 | 1.40 |
ENST00000264775.5 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr12_-_102455846 | 1.38 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr1_-_236228417 | 1.37 |
ENST00000264187.6 |
NID1 |
nidogen 1 |
| chr15_+_40697988 | 1.34 |
ENST00000487418.2 ENST00000479013.2 |
IVD |
isovaleryl-CoA dehydrogenase |
| chr1_-_236228403 | 1.33 |
ENST00000366595.3 |
NID1 |
nidogen 1 |
| chr6_+_30457244 | 1.27 |
ENST00000376630.4 |
HLA-E |
major histocompatibility complex, class I, E |
| chr10_+_13142225 | 1.24 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr14_+_74034310 | 1.23 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr2_+_220143989 | 1.23 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr14_+_104182105 | 1.20 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr13_+_37006421 | 1.19 |
ENST00000255465.4 |
CCNA1 |
cyclin A1 |
| chr6_+_148663729 | 1.18 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr6_+_29691198 | 1.17 |
ENST00000440587.2 ENST00000434407.2 |
HLA-F |
major histocompatibility complex, class I, F |
| chr14_+_104182061 | 1.17 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr8_-_13134045 | 1.05 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr13_+_37005967 | 1.04 |
ENST00000440264.1 ENST00000449823.1 |
CCNA1 |
cyclin A1 |
| chr2_+_220144052 | 1.03 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr13_+_37006398 | 1.03 |
ENST00000418263.1 |
CCNA1 |
cyclin A1 |
| chr18_+_61143994 | 1.01 |
ENST00000382771.4 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr6_+_29691056 | 0.98 |
ENST00000414333.1 ENST00000334668.4 ENST00000259951.7 |
HLA-F |
major histocompatibility complex, class I, F |
| chr12_-_21654479 | 0.96 |
ENST00000421138.2 ENST00000444129.2 ENST00000539672.1 ENST00000542432.1 ENST00000536964.1 ENST00000536240.1 ENST00000396093.3 ENST00000314748.6 |
RECQL |
RecQ protein-like (DNA helicase Q1-like) |
| chr11_-_102651343 | 0.96 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr17_+_42148097 | 0.95 |
ENST00000269097.4 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
| chr16_-_66584059 | 0.92 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr12_+_21654714 | 0.89 |
ENST00000542038.1 ENST00000540141.1 ENST00000229314.5 |
GOLT1B |
golgi transport 1B |
| chr11_+_77532233 | 0.89 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr3_-_105588231 | 0.89 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr11_+_76156045 | 0.87 |
ENST00000533988.1 ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30 |
chromosome 11 open reading frame 30 |
| chr14_+_76127529 | 0.87 |
ENST00000556977.1 ENST00000557636.1 ENST00000286650.5 ENST00000298832.9 |
TTLL5 |
tubulin tyrosine ligase-like family, member 5 |
| chr7_+_150929550 | 0.87 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr7_-_86849883 | 0.86 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr7_-_121784285 | 0.78 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr14_-_76127519 | 0.76 |
ENST00000256319.6 |
C14orf1 |
chromosome 14 open reading frame 1 |
| chr13_+_73632897 | 0.71 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr3_-_160823158 | 0.68 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr11_+_77532155 | 0.66 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr19_-_14606900 | 0.62 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr3_-_151034734 | 0.62 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr10_+_71561649 | 0.61 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_-_160823040 | 0.58 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr17_+_41476327 | 0.58 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr16_-_66583701 | 0.52 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr5_+_66300446 | 0.51 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_242089833 | 0.50 |
ENST00000404405.3 ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr19_-_10121144 | 0.50 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr2_-_55276320 | 0.50 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr17_+_72772621 | 0.49 |
ENST00000335464.5 ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104 |
transmembrane protein 104 |
| chr5_+_174151536 | 0.49 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr10_+_71561630 | 0.49 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr17_+_73452695 | 0.49 |
ENST00000582186.1 ENST00000582455.1 ENST00000581252.1 ENST00000579208.1 |
KIAA0195 |
KIAA0195 |
| chr17_+_74733744 | 0.48 |
ENST00000586689.1 ENST00000587661.1 ENST00000593181.1 ENST00000336509.4 ENST00000355954.3 |
MFSD11 |
major facilitator superfamily domain containing 11 |
| chr22_-_39190116 | 0.47 |
ENST00000406622.1 ENST00000216068.4 ENST00000406199.3 |
SUN2 DNAL4 |
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr7_+_100210133 | 0.47 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr6_-_110501200 | 0.46 |
ENST00000392586.1 ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1 |
WAS protein family, member 1 |
| chr6_-_32095968 | 0.46 |
ENST00000375203.3 ENST00000375201.4 |
ATF6B |
activating transcription factor 6 beta |
| chr20_+_18488137 | 0.45 |
ENST00000450074.1 ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
| chr7_+_100209725 | 0.45 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr4_+_74275057 | 0.43 |
ENST00000511370.1 |
ALB |
albumin |
| chr1_-_48866517 | 0.43 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr22_+_41777927 | 0.43 |
ENST00000266304.4 |
TEF |
thyrotrophic embryonic factor |
| chr7_-_99277610 | 0.42 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr7_+_100209979 | 0.39 |
ENST00000493970.1 ENST00000379527.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr6_-_33285505 | 0.39 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr16_+_6533380 | 0.38 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_113558272 | 0.38 |
ENST00000509061.1 ENST00000508577.1 ENST00000513553.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr17_-_7167279 | 0.38 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr11_-_76155618 | 0.37 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr1_+_151735431 | 0.37 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr3_-_105587879 | 0.37 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_-_103749105 | 0.36 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_7694417 | 0.35 |
ENST00000358368.4 ENST00000534844.1 |
XAB2 |
XPA binding protein 2 |
| chr2_-_74618964 | 0.35 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr6_-_82957433 | 0.35 |
ENST00000306270.7 |
IBTK |
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr12_+_7013897 | 0.34 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr2_-_74619152 | 0.33 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr5_+_142149932 | 0.33 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr9_+_34646651 | 0.32 |
ENST00000378842.3 |
GALT |
galactose-1-phosphate uridylyltransferase |
| chr11_-_76155700 | 0.32 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr12_-_110883346 | 0.30 |
ENST00000547365.1 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr14_+_100842735 | 0.30 |
ENST00000554998.1 ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25 |
WD repeat domain 25 |
| chr1_+_44115814 | 0.29 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr9_+_34646624 | 0.29 |
ENST00000450095.2 ENST00000556278.1 |
GALT GALT |
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr6_-_49681235 | 0.28 |
ENST00000339139.4 |
CRISP2 |
cysteine-rich secretory protein 2 |
| chr17_+_41323204 | 0.28 |
ENST00000542611.1 ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chr11_-_18548426 | 0.28 |
ENST00000357193.3 ENST00000536719.1 |
TSG101 |
tumor susceptibility 101 |
| chr17_+_73452545 | 0.28 |
ENST00000314256.7 |
KIAA0195 |
KIAA0195 |
| chr7_-_36764142 | 0.27 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr20_+_35807449 | 0.27 |
ENST00000237530.6 |
RPN2 |
ribophorin II |
| chr15_-_77712477 | 0.27 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr12_+_7014126 | 0.27 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr4_-_103749313 | 0.27 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr14_-_99947168 | 0.26 |
ENST00000331768.5 |
SETD3 |
SET domain containing 3 |
| chr5_-_82969405 | 0.26 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr1_-_43833628 | 0.26 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr10_-_99447024 | 0.25 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr11_+_33061543 | 0.25 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr10_+_71561704 | 0.24 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr20_+_35807512 | 0.24 |
ENST00000373622.5 |
RPN2 |
ribophorin II |
| chr16_+_6533729 | 0.24 |
ENST00000551752.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_-_121843985 | 0.23 |
ENST00000264808.3 ENST00000428209.2 ENST00000515109.1 ENST00000394435.2 |
PRDM5 |
PR domain containing 5 |
| chr12_+_7014064 | 0.23 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chrY_+_14813160 | 0.23 |
ENST00000338981.3 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
| chr18_+_616672 | 0.22 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr6_+_39760129 | 0.22 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chrX_+_100353153 | 0.22 |
ENST00000423383.1 ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI |
centromere protein I |
| chr7_+_142458507 | 0.22 |
ENST00000492062.1 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr11_+_17298297 | 0.22 |
ENST00000529010.1 |
NUCB2 |
nucleobindin 2 |
| chr6_-_27880174 | 0.22 |
ENST00000303324.2 |
OR2B2 |
olfactory receptor, family 2, subfamily B, member 2 |
| chr17_-_4871085 | 0.22 |
ENST00000575142.1 ENST00000206020.3 |
SPAG7 |
sperm associated antigen 7 |
| chr3_+_40498783 | 0.21 |
ENST00000338970.6 ENST00000396203.2 ENST00000416518.1 |
RPL14 |
ribosomal protein L14 |
| chr4_+_89513574 | 0.21 |
ENST00000402738.1 ENST00000431413.1 ENST00000422770.1 ENST00000407637.1 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr14_-_100842588 | 0.21 |
ENST00000556645.1 ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS |
tryptophanyl-tRNA synthetase |
| chr14_+_22293618 | 0.21 |
ENST00000390432.2 |
TRAV10 |
T cell receptor alpha variable 10 |
| chr3_+_172468505 | 0.20 |
ENST00000427830.1 ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr2_+_113763031 | 0.19 |
ENST00000259211.6 |
IL36A |
interleukin 36, alpha |
| chr20_+_54967409 | 0.19 |
ENST00000415828.1 ENST00000217109.4 ENST00000428552.1 |
CSTF1 |
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr12_-_56753858 | 0.19 |
ENST00000314128.4 ENST00000557235.1 ENST00000418572.2 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
| chr13_+_98086445 | 0.18 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr12_+_27677085 | 0.18 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr11_-_128894053 | 0.17 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr3_+_133292574 | 0.16 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr10_-_104001231 | 0.16 |
ENST00000370002.3 |
PITX3 |
paired-like homeodomain 3 |
| chr11_+_17298255 | 0.15 |
ENST00000531172.1 ENST00000533738.2 ENST00000323688.6 |
NUCB2 |
nucleobindin 2 |
| chr4_+_113558612 | 0.15 |
ENST00000505034.1 ENST00000324052.6 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr8_+_9413410 | 0.15 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr13_-_46626847 | 0.15 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr5_+_142149955 | 0.15 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr3_+_172468472 | 0.14 |
ENST00000232458.5 ENST00000392692.3 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr6_-_29396243 | 0.13 |
ENST00000377148.1 |
OR11A1 |
olfactory receptor, family 11, subfamily A, member 1 |
| chr5_-_112630598 | 0.13 |
ENST00000302475.4 |
MCC |
mutated in colorectal cancers |
| chr1_+_236958554 | 0.13 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr14_+_99947715 | 0.12 |
ENST00000389879.5 ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK |
cyclin K |
| chr9_+_71944241 | 0.12 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr9_-_95640218 | 0.12 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr12_-_9760482 | 0.11 |
ENST00000229402.3 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
| chr12_-_10955226 | 0.10 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr19_-_12833361 | 0.10 |
ENST00000592287.1 |
TNPO2 |
transportin 2 |
| chr22_-_17640110 | 0.10 |
ENST00000399852.3 ENST00000336737.4 |
CECR5 |
cat eye syndrome chromosome region, candidate 5 |
| chr7_-_72971934 | 0.10 |
ENST00000411832.1 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr10_+_7830092 | 0.09 |
ENST00000356708.7 |
ATP5C1 |
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr3_+_158519654 | 0.09 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr19_-_19302931 | 0.09 |
ENST00000444486.3 ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B MEF2BNB MEF2B |
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr9_+_130565147 | 0.09 |
ENST00000373247.2 ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS |
folylpolyglutamate synthase |
| chr21_-_38639813 | 0.09 |
ENST00000309117.6 ENST00000398998.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr6_-_31125850 | 0.09 |
ENST00000507751.1 ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1 |
coiled-coil alpha-helical rod protein 1 |
| chr6_-_155776966 | 0.08 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr5_-_137878887 | 0.07 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr1_-_204380919 | 0.07 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr1_+_28261621 | 0.07 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr6_+_26402465 | 0.07 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr19_-_41870026 | 0.06 |
ENST00000243578.3 |
B9D2 |
B9 protein domain 2 |
| chr17_-_77770830 | 0.06 |
ENST00000269385.4 |
CBX8 |
chromobox homolog 8 |
| chr7_-_87342564 | 0.06 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr4_-_103748880 | 0.06 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_66511540 | 0.05 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr22_-_42017021 | 0.05 |
ENST00000263256.6 |
DESI1 |
desumoylating isopeptidase 1 |
| chr1_-_13390765 | 0.05 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr5_-_94890648 | 0.05 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr1_-_13673511 | 0.05 |
ENST00000344998.3 ENST00000334600.6 |
PRAMEF14 |
PRAME family member 14 |
| chr6_+_29624758 | 0.04 |
ENST00000376917.3 ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr11_+_67798114 | 0.04 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr14_+_52456327 | 0.04 |
ENST00000556760.1 |
C14orf166 |
chromosome 14 open reading frame 166 |
| chr22_+_42017459 | 0.04 |
ENST00000405878.1 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chrX_+_113818545 | 0.04 |
ENST00000371951.1 ENST00000276198.1 ENST00000371950.3 |
HTR2C |
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| chr6_-_62996066 | 0.04 |
ENST00000281156.4 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
| chr6_+_52051171 | 0.04 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr5_-_137548997 | 0.04 |
ENST00000505120.1 ENST00000394886.2 ENST00000394884.3 |
CDC23 |
cell division cycle 23 |
| chr11_-_133826852 | 0.03 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr11_+_67798090 | 0.03 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr10_-_48416849 | 0.03 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr6_-_29395509 | 0.02 |
ENST00000377147.2 |
OR11A1 |
olfactory receptor, family 11, subfamily A, member 1 |
| chr17_-_40828969 | 0.02 |
ENST00000591022.1 ENST00000587627.1 ENST00000293349.6 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_-_41466555 | 0.02 |
ENST00000586231.1 |
LINC00910 |
long intergenic non-protein coding RNA 910 |
| chr11_+_7110165 | 0.02 |
ENST00000306904.5 |
RBMXL2 |
RNA binding motif protein, X-linked-like 2 |
| chr6_+_160542821 | 0.02 |
ENST00000366963.4 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr22_+_42017280 | 0.01 |
ENST00000402580.3 ENST00000428575.2 ENST00000359308.4 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr16_-_55866997 | 0.01 |
ENST00000360526.3 ENST00000361503.4 |
CES1 |
carboxylesterase 1 |
| chr14_-_104181771 | 0.01 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr18_+_5238055 | 0.01 |
ENST00000582363.1 ENST00000582008.1 ENST00000580082.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr1_+_28261492 | 0.01 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr3_+_67705121 | 0.01 |
ENST00000464420.1 ENST00000482677.1 |
RP11-81N13.1 |
RP11-81N13.1 |
| chr6_+_160542870 | 0.00 |
ENST00000324965.4 ENST00000457470.2 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr12_-_95611149 | 0.00 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_12902289 | 0.00 |
ENST00000302754.4 |
JUNB |
jun B proto-oncogene |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.5 | 2.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.4 | 1.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.4 | 2.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.4 | 2.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.3 | 0.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.9 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 4.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.9 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 6.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 3.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 3.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 7.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 4.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 3.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.6 | 2.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.5 | 3.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 1.3 | GO:0032679 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) protection from natural killer cell mediated cytotoxicity(GO:0042270) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.4 | 1.4 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 0.8 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 2.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 7.0 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.2 | 0.6 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.5 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.2 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.8 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 1.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 3.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 2.5 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 3.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 2.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 2.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 1.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.3 | GO:1900113 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.9 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.5 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.4 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.3 | 1.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 3.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 4.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 4.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 3.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 5.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |