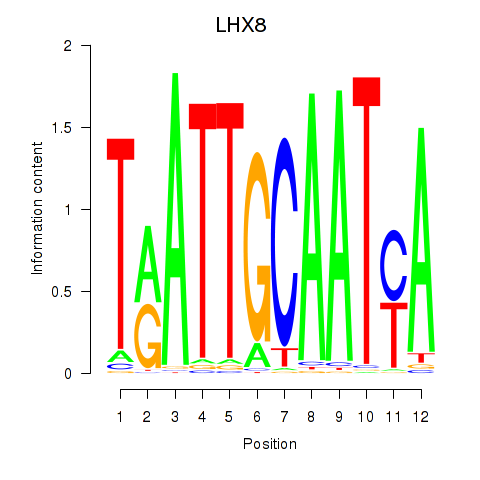
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for LHX8
Z-value: 0.79
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LHX8 |
Activity profile of LHX8 motif
Sorted Z-values of LHX8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74275057 | 2.58 |
ENST00000511370.1 |
ALB |
albumin |
| chr4_+_166300084 | 1.96 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr4_-_110723134 | 1.67 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr4_-_110723335 | 1.65 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr4_-_110723194 | 1.64 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr7_-_22234381 | 1.58 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr10_-_101825151 | 1.16 |
ENST00000441382.1 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr2_-_158345462 | 1.06 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr12_-_8043736 | 0.96 |
ENST00000539924.1 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr1_+_70876926 | 0.71 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr12_-_8088871 | 0.70 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr2_+_201994208 | 0.67 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chrX_-_20236970 | 0.64 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr4_+_155484155 | 0.57 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr9_-_5833027 | 0.56 |
ENST00000339450.5 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
| chr7_+_74188309 | 0.55 |
ENST00000289473.4 ENST00000433458.1 |
NCF1 |
neutrophil cytosolic factor 1 |
| chr8_-_82395461 | 0.55 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr7_+_142498725 | 0.54 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr7_-_137028498 | 0.51 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr3_+_173116225 | 0.50 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr12_-_15038779 | 0.48 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr7_-_137028534 | 0.46 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr3_-_57199397 | 0.46 |
ENST00000296318.7 |
IL17RD |
interleukin 17 receptor D |
| chr10_+_94352956 | 0.41 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr2_+_201994042 | 0.41 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chrX_+_108779004 | 0.40 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chrX_+_84258832 | 0.39 |
ENST00000373173.2 |
APOOL |
apolipoprotein O-like |
| chr4_+_155484103 | 0.39 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr9_-_36276966 | 0.39 |
ENST00000543356.2 ENST00000396594.3 |
GNE |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr19_-_4535233 | 0.39 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr1_+_28696111 | 0.38 |
ENST00000373839.3 |
PHACTR4 |
phosphatase and actin regulator 4 |
| chr2_+_86947296 | 0.37 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr15_-_65426174 | 0.37 |
ENST00000204549.4 |
PDCD7 |
programmed cell death 7 |
| chr9_-_20622478 | 0.36 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr9_-_123676827 | 0.36 |
ENST00000546084.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr4_-_83812402 | 0.33 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr2_-_88285309 | 0.33 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr7_+_55980331 | 0.32 |
ENST00000429591.2 |
ZNF713 |
zinc finger protein 713 |
| chr4_+_68424434 | 0.31 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr1_-_212004090 | 0.31 |
ENST00000366997.4 |
LPGAT1 |
lysophosphatidylglycerol acyltransferase 1 |
| chr3_-_119182523 | 0.30 |
ENST00000319172.5 |
TMEM39A |
transmembrane protein 39A |
| chr4_-_186733363 | 0.30 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_83812248 | 0.30 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr19_+_34855925 | 0.30 |
ENST00000590375.1 ENST00000356487.5 |
GPI |
glucose-6-phosphate isomerase |
| chr7_-_144435985 | 0.29 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr1_+_212208919 | 0.29 |
ENST00000366991.4 ENST00000542077.1 |
DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr2_+_87144738 | 0.29 |
ENST00000559485.1 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr1_-_236445251 | 0.29 |
ENST00000354619.5 ENST00000327333.8 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
| chr1_-_160492994 | 0.28 |
ENST00000368055.1 ENST00000368057.3 ENST00000368059.3 |
SLAMF6 |
SLAM family member 6 |
| chr19_+_34855874 | 0.28 |
ENST00000588991.2 |
GPI |
glucose-6-phosphate isomerase |
| chr15_-_34880646 | 0.28 |
ENST00000543376.1 |
GOLGA8A |
golgin A8 family, member A |
| chr2_-_50201327 | 0.28 |
ENST00000412315.1 |
NRXN1 |
neurexin 1 |
| chr2_-_152830441 | 0.28 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_175511859 | 0.27 |
ENST00000503724.2 ENST00000253490.4 |
FAM153B |
family with sequence similarity 153, member B |
| chr14_+_39583427 | 0.27 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr22_-_29107919 | 0.26 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr12_+_9822331 | 0.25 |
ENST00000545918.1 ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D |
C-type lectin domain family 2, member D |
| chr2_-_136288113 | 0.25 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr2_+_87135076 | 0.25 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr20_+_34824355 | 0.24 |
ENST00000397286.3 ENST00000320849.4 ENST00000373932.3 |
AAR2 |
AAR2 splicing factor homolog (S. cerevisiae) |
| chr3_-_52804872 | 0.24 |
ENST00000535191.1 ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4 |
NIMA-related kinase 4 |
| chr19_+_12175504 | 0.23 |
ENST00000439326.3 |
ZNF844 |
zinc finger protein 844 |
| chr2_-_152118352 | 0.23 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chrX_+_108779870 | 0.22 |
ENST00000372107.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr12_-_71551868 | 0.22 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr5_-_147162263 | 0.22 |
ENST00000333010.6 ENST00000265272.5 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr17_+_58018269 | 0.21 |
ENST00000591035.1 |
RP11-178C3.1 |
Uncharacterized protein |
| chr2_-_136678123 | 0.20 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr22_-_50219548 | 0.20 |
ENST00000404034.1 |
BRD1 |
bromodomain containing 1 |
| chr5_-_24645078 | 0.20 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr2_-_152830479 | 0.20 |
ENST00000360283.6 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_+_95616933 | 0.20 |
ENST00000604203.1 |
RP11-57H12.6 |
TMEM56-RWDD3 readthrough |
| chrX_+_73164149 | 0.19 |
ENST00000602938.1 ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX |
JPX transcript, XIST activator (non-protein coding) |
| chr14_+_23067146 | 0.19 |
ENST00000428304.2 |
ABHD4 |
abhydrolase domain containing 4 |
| chr11_-_72504637 | 0.19 |
ENST00000536377.1 ENST00000359373.5 |
STARD10 ARAP1 |
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_-_225266743 | 0.19 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr3_+_195447738 | 0.19 |
ENST00000447234.2 ENST00000320736.6 ENST00000436408.1 |
MUC20 |
mucin 20, cell surface associated |
| chr2_-_225266711 | 0.19 |
ENST00000389874.3 |
FAM124B |
family with sequence similarity 124B |
| chr4_+_106631966 | 0.18 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr15_+_65843130 | 0.17 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_+_148545586 | 0.17 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr3_-_149470229 | 0.17 |
ENST00000473414.1 |
COMMD2 |
COMM domain containing 2 |
| chr19_+_44716678 | 0.16 |
ENST00000586228.1 ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227 |
zinc finger protein 227 |
| chr4_+_57302297 | 0.16 |
ENST00000399688.3 ENST00000512576.1 |
PAICS |
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr9_-_21166659 | 0.16 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
| chr7_-_20256965 | 0.16 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr12_-_53893399 | 0.16 |
ENST00000267079.2 |
MAP3K12 |
mitogen-activated protein kinase kinase kinase 12 |
| chr13_+_53216565 | 0.16 |
ENST00000357495.2 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr10_+_135207598 | 0.16 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr13_+_98605902 | 0.15 |
ENST00000460070.1 ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5 |
importin 5 |
| chr8_+_119294456 | 0.15 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr6_-_64029879 | 0.14 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr10_+_135207623 | 0.14 |
ENST00000317502.6 ENST00000432508.3 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr4_-_100242549 | 0.14 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr2_+_234826016 | 0.13 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_-_42144949 | 0.12 |
ENST00000591247.1 |
LSM12 |
LSM12 homolog (S. cerevisiae) |
| chr12_-_88974236 | 0.12 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chrX_+_103031421 | 0.12 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr4_-_109684120 | 0.12 |
ENST00000512646.1 ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL |
ethanolamine-phosphate phospho-lyase |
| chr12_-_71551652 | 0.12 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr1_+_45241109 | 0.11 |
ENST00000396651.3 |
RPS8 |
ribosomal protein S8 |
| chr15_-_34610962 | 0.11 |
ENST00000290209.5 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr2_+_185463093 | 0.11 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr4_-_48082192 | 0.11 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr2_-_74618964 | 0.11 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr5_+_140235469 | 0.11 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr15_-_35047166 | 0.11 |
ENST00000290374.4 |
GJD2 |
gap junction protein, delta 2, 36kDa |
| chr2_-_74619152 | 0.10 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr6_+_26251835 | 0.10 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr14_+_23012122 | 0.10 |
ENST00000390534.1 |
TRAJ3 |
T cell receptor alpha joining 3 |
| chr10_-_70231639 | 0.10 |
ENST00000551118.2 ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2 |
DNA replication helicase/nuclease 2 |
| chr11_+_72975559 | 0.09 |
ENST00000349767.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_+_115862858 | 0.09 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr7_-_140482926 | 0.09 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr11_+_72975524 | 0.08 |
ENST00000540342.1 ENST00000542092.1 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr4_-_186125077 | 0.08 |
ENST00000458385.2 ENST00000514798.1 ENST00000296775.6 |
KIAA1430 |
KIAA1430 |
| chr6_-_155776966 | 0.08 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr12_+_59989918 | 0.08 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_+_72975578 | 0.08 |
ENST00000393592.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr8_+_75262612 | 0.08 |
ENST00000220822.7 |
GDAP1 |
ganglioside induced differentiation associated protein 1 |
| chr1_+_119957554 | 0.07 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr16_-_20339123 | 0.07 |
ENST00000381360.5 |
GP2 |
glycoprotein 2 (zymogen granule membrane) |
| chr19_+_21264980 | 0.07 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr2_+_108145913 | 0.07 |
ENST00000443205.1 |
AC096669.3 |
AC096669.3 |
| chr19_+_47852538 | 0.06 |
ENST00000328771.4 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr14_+_102829300 | 0.06 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr8_+_17354617 | 0.06 |
ENST00000470360.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr14_+_72399833 | 0.06 |
ENST00000553530.1 ENST00000556437.1 |
RGS6 |
regulator of G-protein signaling 6 |
| chr8_+_27168988 | 0.06 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr12_+_101988774 | 0.05 |
ENST00000545503.2 ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr7_-_120498357 | 0.05 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr4_-_153332886 | 0.05 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr21_-_34863998 | 0.05 |
ENST00000402202.1 ENST00000381947.3 |
DNAJC28 |
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr12_+_101988627 | 0.05 |
ENST00000547405.1 ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr16_-_15149917 | 0.05 |
ENST00000287706.3 |
NTAN1 |
N-terminal asparagine amidase |
| chr16_-_15149828 | 0.05 |
ENST00000566419.1 ENST00000568320.1 |
NTAN1 |
N-terminal asparagine amidase |
| chr17_-_76719807 | 0.05 |
ENST00000589297.1 |
CYTH1 |
cytohesin 1 |
| chr11_-_66964638 | 0.04 |
ENST00000444002.2 |
AP001885.1 |
AP001885.1 |
| chr19_-_48547294 | 0.04 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chr11_-_84634217 | 0.04 |
ENST00000524982.1 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr1_+_53662101 | 0.04 |
ENST00000371486.3 |
CPT2 |
carnitine palmitoyltransferase 2 |
| chr4_-_68749699 | 0.04 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr6_+_153071925 | 0.03 |
ENST00000367244.3 ENST00000367243.3 |
VIP |
vasoactive intestinal peptide |
| chr4_-_68749745 | 0.03 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr4_+_71108300 | 0.03 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr3_-_48956818 | 0.03 |
ENST00000408959.2 |
ARIH2OS |
ariadne homolog 2 opposite strand |
| chr12_-_11548496 | 0.03 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr6_-_24489842 | 0.03 |
ENST00000230036.1 |
GPLD1 |
glycosylphosphatidylinositol specific phospholipase D1 |
| chr9_-_21217310 | 0.03 |
ENST00000380216.1 |
IFNA16 |
interferon, alpha 16 |
| chr6_+_32006042 | 0.02 |
ENST00000418967.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_+_71248795 | 0.02 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr12_-_21487829 | 0.02 |
ENST00000445053.1 ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2 |
solute carrier organic anion transporter family, member 1A2 |
| chr3_-_47934234 | 0.02 |
ENST00000420772.2 |
MAP4 |
microtubule-associated protein 4 |
| chr8_+_17354587 | 0.01 |
ENST00000494857.1 ENST00000522656.1 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr1_+_113009163 | 0.01 |
ENST00000256640.5 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
| chr12_+_14572070 | 0.01 |
ENST00000545769.1 ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr19_-_9006766 | 0.01 |
ENST00000599436.1 |
MUC16 |
mucin 16, cell surface associated |
| chr8_+_22424551 | 0.01 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr16_-_2314222 | 0.01 |
ENST00000566397.1 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
| chr1_+_144989309 | 0.01 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr1_-_217262933 | 0.01 |
ENST00000359162.2 |
ESRRG |
estrogen-related receptor gamma |
| chr13_+_96085847 | 0.01 |
ENST00000376873.3 |
CLDN10 |
claudin 10 |
| chr5_+_140165876 | 0.00 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr6_+_26087509 | 0.00 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr8_-_141810634 | 0.00 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr5_+_157158205 | 0.00 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 3.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.0 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.2 | 0.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.5 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.4 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 1.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 1.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 5.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1903401 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine transmembrane transport(GO:1903401) L-lysine import into cell(GO:1903410) |
| 0.0 | 1.1 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.6 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.3 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 3.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.6 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.2 | 2.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 5.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |