Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
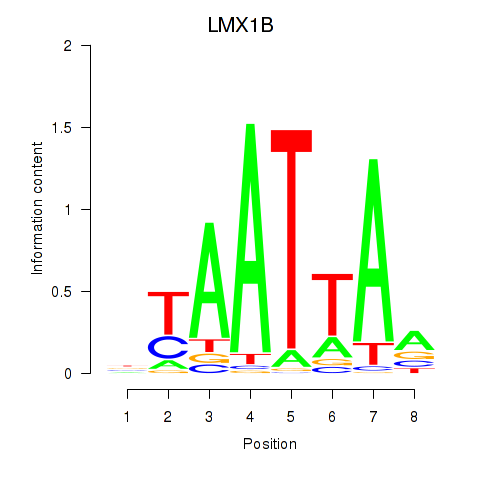
Results for LMX1B_MNX1_RAX2
Z-value: 0.84
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LMX1B
|
ENSG00000136944.13 | LMX1B |
|
MNX1
|
ENSG00000130675.10 | MNX1 |
|
RAX2
|
ENSG00000173976.11 | RAX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MNX1 | hg19_v2_chr7_-_156803329_156803362 | 0.34 | 1.9e-01 | Click! |
| LMX1B | hg19_v2_chr9_+_129376722_129376748 | 0.17 | 5.2e-01 | Click! |
| RAX2 | hg19_v2_chr19_-_3772209_3772236 | 0.09 | 7.4e-01 | Click! |
Activity profile of LMX1B_MNX1_RAX2 motif
Sorted Z-values of LMX1B_MNX1_RAX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LMX1B_MNX1_RAX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_169418195 | 4.78 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr12_-_28123206 | 3.38 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr17_-_64225508 | 3.08 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_28122980 | 2.87 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_52967600 | 2.79 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr7_+_134464414 | 2.36 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr4_+_169418255 | 2.27 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_-_151034734 | 2.21 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr11_-_129062093 | 1.97 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr7_+_134464376 | 1.91 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr12_-_89746173 | 1.86 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr1_-_237167718 | 1.80 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chrX_-_13835147 | 1.61 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr7_-_107642348 | 1.60 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr16_-_29910853 | 1.50 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chrX_+_105937068 | 1.38 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_-_51522955 | 1.26 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chrX_-_106243451 | 1.18 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr2_+_68961934 | 1.11 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 1.10 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr7_-_27169801 | 1.09 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr3_-_79816965 | 0.97 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_+_66300446 | 0.95 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_143226979 | 0.88 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr5_+_174151536 | 0.86 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr17_-_77924627 | 0.83 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chrX_-_10851762 | 0.80 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr4_-_143227088 | 0.79 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr18_+_29027696 | 0.70 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr17_+_72427477 | 0.67 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr5_-_1882858 | 0.64 |
ENST00000511126.1 ENST00000231357.2 |
IRX4 |
iroquois homeobox 4 |
| chr7_-_87342564 | 0.63 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr20_+_30697298 | 0.63 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr3_+_111717600 | 0.62 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr2_+_187454749 | 0.61 |
ENST00000261023.3 ENST00000374907.3 |
ITGAV |
integrin, alpha V |
| chr1_+_68150744 | 0.60 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr2_+_68962014 | 0.59 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr8_-_18744528 | 0.58 |
ENST00000523619.1 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr3_+_111718036 | 0.56 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr12_+_81110684 | 0.55 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr8_+_105235572 | 0.53 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr2_+_234826016 | 0.52 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_+_47448102 | 0.51 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr2_-_208031943 | 0.51 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_149293990 | 0.49 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr8_+_101170563 | 0.49 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr3_+_111717511 | 0.48 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr1_+_225600404 | 0.47 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr4_-_186696425 | 0.46 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr15_+_58702742 | 0.46 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr14_+_21156915 | 0.45 |
ENST00000397990.4 ENST00000555597.1 |
ANG RNASE4 |
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr1_-_227505289 | 0.43 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr19_-_51523412 | 0.42 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr5_+_125758865 | 0.42 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr19_-_51523275 | 0.42 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr2_-_207024233 | 0.41 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_+_125758813 | 0.39 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr1_-_24469602 | 0.38 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr6_-_87804815 | 0.38 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr19_+_50016610 | 0.37 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_-_68698222 | 0.35 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_+_180601139 | 0.32 |
ENST00000367590.4 ENST00000367589.3 |
XPR1 |
xenotropic and polytropic retrovirus receptor 1 |
| chr3_+_111718173 | 0.32 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr3_+_158787041 | 0.31 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr6_+_151646800 | 0.31 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr19_+_50016411 | 0.30 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_-_39191107 | 0.30 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr1_-_94586651 | 0.30 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr18_-_33709268 | 0.29 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr1_+_214161272 | 0.29 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr16_-_4852915 | 0.29 |
ENST00000322048.7 |
ROGDI |
rogdi homolog (Drosophila) |
| chr3_-_178984759 | 0.29 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr7_-_22234381 | 0.29 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_38064198 | 0.29 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr1_+_214161854 | 0.28 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr16_+_69345243 | 0.27 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr17_+_41363854 | 0.26 |
ENST00000588693.1 ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A |
transmembrane protein 106A |
| chr7_+_100136811 | 0.24 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr2_+_109237717 | 0.23 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr14_+_104182061 | 0.23 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr7_-_107443652 | 0.22 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr2_-_216946500 | 0.22 |
ENST00000265322.7 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
| chr14_+_104182105 | 0.21 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr7_-_143599207 | 0.19 |
ENST00000355951.2 ENST00000479870.1 ENST00000478172.1 |
FAM115A |
family with sequence similarity 115, member A |
| chr15_+_93443419 | 0.18 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr2_-_74619152 | 0.18 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chrX_+_78003204 | 0.18 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr2_-_207024134 | 0.18 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr18_+_46065393 | 0.18 |
ENST00000256413.3 |
CTIF |
CBP80/20-dependent translation initiation factor |
| chr1_+_210501589 | 0.18 |
ENST00000413764.2 ENST00000541565.1 |
HHAT |
hedgehog acyltransferase |
| chr12_-_10978957 | 0.17 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr6_-_136788001 | 0.17 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr2_+_171034646 | 0.15 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr3_+_130569429 | 0.15 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_-_92952433 | 0.14 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr4_+_69313145 | 0.14 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr6_-_111927449 | 0.14 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr7_+_138145076 | 0.14 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr4_+_175204818 | 0.14 |
ENST00000503780.1 |
CEP44 |
centrosomal protein 44kDa |
| chr17_-_72772462 | 0.14 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_+_63606373 | 0.13 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr1_+_117544366 | 0.13 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr7_-_83278322 | 0.13 |
ENST00000307792.3 |
SEMA3E |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr7_-_44580861 | 0.13 |
ENST00000546276.1 ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1 |
NPC1-like 1 |
| chr5_-_24645078 | 0.13 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr4_+_3388057 | 0.13 |
ENST00000538395.1 |
RGS12 |
regulator of G-protein signaling 12 |
| chr19_-_5903714 | 0.13 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr16_+_68279207 | 0.13 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr2_+_170440844 | 0.12 |
ENST00000260970.3 ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr14_-_25479811 | 0.12 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr4_+_71457970 | 0.11 |
ENST00000322937.6 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr12_+_26348582 | 0.11 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr4_+_88754113 | 0.11 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr1_-_242162375 | 0.11 |
ENST00000357246.3 |
MAP1LC3C |
microtubule-associated protein 1 light chain 3 gamma |
| chr2_-_207023918 | 0.11 |
ENST00000455934.2 ENST00000449699.1 ENST00000454195.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_+_177015122 | 0.11 |
ENST00000468418.3 |
HOXD3 |
homeobox D3 |
| chr19_-_14064114 | 0.11 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chr19_-_46088068 | 0.11 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr1_-_68698197 | 0.10 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr3_-_52090461 | 0.10 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr19_-_36304201 | 0.10 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr12_-_118796910 | 0.10 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr4_+_41614720 | 0.10 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr11_+_92702886 | 0.10 |
ENST00000257068.2 ENST00000528076.1 |
MTNR1B |
melatonin receptor 1B |
| chr14_+_32798547 | 0.10 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr17_-_39646116 | 0.10 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr13_+_53602894 | 0.10 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr4_+_71458012 | 0.10 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr1_+_62439037 | 0.10 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr6_-_136847099 | 0.09 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr19_+_13049413 | 0.09 |
ENST00000316448.5 ENST00000588454.1 |
CALR |
calreticulin |
| chr16_+_57279004 | 0.09 |
ENST00000219204.3 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr13_-_44735393 | 0.09 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr20_-_21494654 | 0.09 |
ENST00000377142.4 |
NKX2-2 |
NK2 homeobox 2 |
| chr13_-_95131923 | 0.09 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chr2_+_45168875 | 0.09 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr1_+_154401791 | 0.09 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chr12_+_4385230 | 0.09 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr2_-_190927447 | 0.08 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr3_-_157824292 | 0.08 |
ENST00000483851.2 |
SHOX2 |
short stature homeobox 2 |
| chr4_-_139163491 | 0.08 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr17_-_38859996 | 0.08 |
ENST00000264651.2 |
KRT24 |
keratin 24 |
| chrX_-_18690210 | 0.08 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr4_+_88754069 | 0.08 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chrX_+_135730297 | 0.08 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chrX_+_144908928 | 0.08 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr8_+_117950422 | 0.07 |
ENST00000378279.3 |
AARD |
alanine and arginine rich domain containing protein |
| chr13_-_36050819 | 0.07 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr12_-_16759711 | 0.07 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr14_+_72052983 | 0.07 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr12_+_41831485 | 0.07 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr1_+_50569575 | 0.07 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr12_-_53994805 | 0.07 |
ENST00000328463.7 |
ATF7 |
activating transcription factor 7 |
| chr3_-_114477787 | 0.07 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_-_86650045 | 0.06 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chrX_-_19988382 | 0.06 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr9_-_136933134 | 0.06 |
ENST00000303407.7 |
BRD3 |
bromodomain containing 3 |
| chrX_+_107288239 | 0.06 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr14_-_78083112 | 0.06 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr10_+_102222798 | 0.06 |
ENST00000343737.5 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
| chr15_+_44092784 | 0.06 |
ENST00000458412.1 |
HYPK |
huntingtin interacting protein K |
| chr6_-_31107127 | 0.06 |
ENST00000259845.4 |
PSORS1C2 |
psoriasis susceptibility 1 candidate 2 |
| chr3_-_167191814 | 0.06 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr16_+_89334512 | 0.06 |
ENST00000602042.1 |
AC137932.1 |
AC137932.1 |
| chr8_-_93107696 | 0.05 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_+_122688090 | 0.05 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr2_-_100987007 | 0.05 |
ENST00000595083.1 |
AC012493.2 |
Uncharacterized protein |
| chr22_-_40929812 | 0.05 |
ENST00000422851.1 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
| chr12_-_86650077 | 0.05 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_+_26505179 | 0.05 |
ENST00000376261.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr9_+_82188077 | 0.05 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_48937821 | 0.05 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr16_+_30669720 | 0.05 |
ENST00000356166.6 |
FBRS |
fibrosin |
| chr4_+_88571429 | 0.05 |
ENST00000339673.6 ENST00000282479.7 |
DMP1 |
dentin matrix acidic phosphoprotein 1 |
| chr6_-_167797887 | 0.05 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr2_+_207024306 | 0.05 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chrX_+_135730373 | 0.04 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr21_-_31538971 | 0.04 |
ENST00000286808.3 |
CLDN17 |
claudin 17 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr17_-_56296580 | 0.04 |
ENST00000313863.6 ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1 |
Meckel syndrome, type 1 |
| chr1_-_149900122 | 0.04 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr12_-_53171128 | 0.04 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr4_+_41614909 | 0.03 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr10_-_96829246 | 0.03 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_-_48937838 | 0.03 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr6_+_153071925 | 0.03 |
ENST00000367244.3 ENST00000367243.3 |
VIP |
vasoactive intestinal peptide |
| chr5_-_138534071 | 0.03 |
ENST00000394817.2 |
SIL1 |
SIL1 nucleotide exchange factor |
| chr3_+_155838337 | 0.03 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_+_22050546 | 0.03 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr1_-_151762943 | 0.03 |
ENST00000368825.3 ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH |
tudor and KH domain containing |
| chr19_+_4402659 | 0.03 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr2_-_209010874 | 0.03 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr1_-_204135450 | 0.03 |
ENST00000272190.8 ENST00000367195.2 |
REN |
renin |
| chr13_-_88323218 | 0.02 |
ENST00000436290.2 ENST00000453832.2 ENST00000606590.1 |
MIR4500HG |
MIR4500 host gene (non-protein coding) |
| chr12_-_10955226 | 0.02 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr4_+_88529681 | 0.02 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr8_-_42234745 | 0.02 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr6_-_136847610 | 0.02 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr10_-_115904361 | 0.02 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.4 | 1.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.3 | 7.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 0.9 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.3 | 3.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 6.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 0.6 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.2 | 1.6 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.6 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 1.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 0.6 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 2.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.7 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 1.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 4.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 6.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 2.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 6.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 6.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 4.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 2.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 5.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 4.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 3.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 4.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |