Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MAFA
Z-value: 0.81
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAFA |
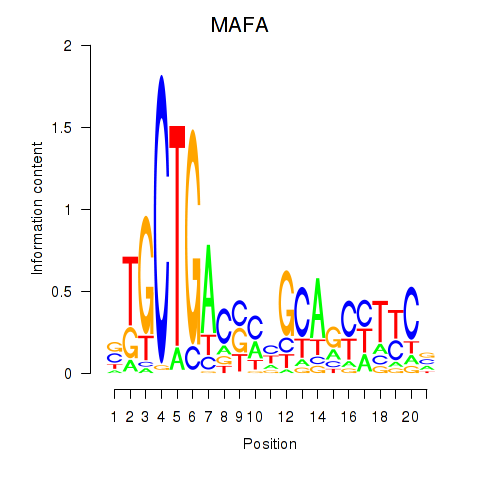
Activity profile of MAFA motif
Sorted Z-values of MAFA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_124221036 | 2.52 |
ENST00000368984.3 |
HTRA1 |
HtrA serine peptidase 1 |
| chr11_-_104817919 | 2.18 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_85811525 | 2.15 |
ENST00000306384.4 |
VAMP5 |
vesicle-associated membrane protein 5 |
| chr1_-_153517473 | 1.99 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr17_-_10325261 | 1.74 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr10_-_79397391 | 1.47 |
ENST00000286628.8 ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_57045228 | 1.41 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr7_-_151433342 | 1.35 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_-_30029574 | 1.34 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr16_+_56598961 | 1.33 |
ENST00000219162.3 |
MT4 |
metallothionein 4 |
| chr10_+_120967072 | 1.24 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr7_-_30029367 | 1.23 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr6_+_116692102 | 1.23 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr6_+_89791507 | 1.21 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr9_+_97488939 | 1.19 |
ENST00000277198.2 ENST00000297979.5 |
C9orf3 |
chromosome 9 open reading frame 3 |
| chr12_-_21654479 | 1.11 |
ENST00000421138.2 ENST00000444129.2 ENST00000539672.1 ENST00000542432.1 ENST00000536964.1 ENST00000536240.1 ENST00000396093.3 ENST00000314748.6 |
RECQL |
RecQ protein-like (DNA helicase Q1-like) |
| chr4_-_22517620 | 1.05 |
ENST00000502482.1 ENST00000334304.5 |
GPR125 |
G protein-coupled receptor 125 |
| chr12_+_79258444 | 0.89 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr1_+_163039143 | 0.87 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr12_+_21654714 | 0.87 |
ENST00000542038.1 ENST00000540141.1 ENST00000229314.5 |
GOLT1B |
golgi transport 1B |
| chr1_-_203144941 | 0.83 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr7_-_86849883 | 0.79 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr2_+_191745535 | 0.74 |
ENST00000320717.3 |
GLS |
glutaminase |
| chr3_+_107241783 | 0.72 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr15_+_80696666 | 0.71 |
ENST00000303329.4 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr12_+_79258547 | 0.62 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr16_-_29517141 | 0.62 |
ENST00000550665.1 |
RP11-231C14.4 |
Uncharacterized protein |
| chr5_-_94620239 | 0.61 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr2_+_118846008 | 0.55 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr12_+_56546363 | 0.53 |
ENST00000551834.1 ENST00000552568.1 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chrX_-_153141302 | 0.53 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chrX_-_92928557 | 0.52 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr4_-_10118348 | 0.47 |
ENST00000502702.1 |
WDR1 |
WD repeat domain 1 |
| chr16_-_21436459 | 0.42 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr6_+_7727030 | 0.38 |
ENST00000283147.6 |
BMP6 |
bone morphogenetic protein 6 |
| chr10_-_79397479 | 0.38 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_-_106959631 | 0.36 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr11_-_66103867 | 0.36 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr14_+_24605389 | 0.34 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr16_+_22524844 | 0.30 |
ENST00000538606.1 ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr16_-_21868739 | 0.30 |
ENST00000415645.2 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr2_+_191745560 | 0.29 |
ENST00000338435.4 |
GLS |
glutaminase |
| chr7_-_100860851 | 0.28 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr11_-_2182388 | 0.28 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr10_-_29811456 | 0.27 |
ENST00000535393.1 |
SVIL |
supervillin |
| chr20_-_23066953 | 0.26 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr9_+_2622085 | 0.25 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr3_-_48754599 | 0.23 |
ENST00000413654.1 ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
| chr16_-_29415350 | 0.23 |
ENST00000524087.1 |
NPIPB11 |
nuclear pore complex interacting protein family, member B11 |
| chr22_-_28197486 | 0.23 |
ENST00000302326.4 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
| chr1_+_90286562 | 0.22 |
ENST00000525774.1 ENST00000337338.5 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr5_-_114505624 | 0.22 |
ENST00000513154.1 |
TRIM36 |
tripartite motif containing 36 |
| chr2_+_33172221 | 0.22 |
ENST00000354476.3 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_102624977 | 0.19 |
ENST00000441002.1 |
IL1R2 |
interleukin 1 receptor, type II |
| chr4_+_2470664 | 0.18 |
ENST00000314289.8 ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4 |
ring finger protein 4 |
| chrY_-_1461617 | 0.17 |
ENSTR0000381401.5 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr11_+_33563821 | 0.16 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr5_-_118324200 | 0.16 |
ENST00000515439.3 ENST00000510708.1 |
DTWD2 |
DTW domain containing 2 |
| chr10_-_102289611 | 0.15 |
ENST00000299166.4 ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8 SEC31B |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr4_+_74606223 | 0.15 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chrX_-_43741594 | 0.15 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr8_+_85095553 | 0.15 |
ENST00000521268.1 |
RALYL |
RALY RNA binding protein-like |
| chr5_+_138629389 | 0.12 |
ENST00000504045.1 ENST00000504311.1 ENST00000502499.1 |
MATR3 |
matrin 3 |
| chr9_+_2621798 | 0.11 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr11_+_72929319 | 0.11 |
ENST00000393597.2 ENST00000311131.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr5_+_138629337 | 0.11 |
ENST00000394805.3 ENST00000512876.1 ENST00000513678.1 |
MATR3 |
matrin 3 |
| chr19_+_12949251 | 0.11 |
ENST00000251472.4 |
MAST1 |
microtubule associated serine/threonine kinase 1 |
| chr8_+_85095497 | 0.11 |
ENST00000522455.1 ENST00000521695.1 |
RALYL |
RALY RNA binding protein-like |
| chr7_-_73133959 | 0.10 |
ENST00000395155.3 ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A |
syntaxin 1A (brain) |
| chr8_+_85095769 | 0.09 |
ENST00000518566.1 |
RALYL |
RALY RNA binding protein-like |
| chr22_+_23161491 | 0.09 |
ENST00000390316.2 |
IGLV3-9 |
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr17_-_42019836 | 0.09 |
ENST00000225992.3 |
PPY |
pancreatic polypeptide |
| chr11_-_66103932 | 0.08 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr18_+_77623668 | 0.08 |
ENST00000316249.3 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
| chrX_-_65859096 | 0.08 |
ENST00000456230.2 |
EDA2R |
ectodysplasin A2 receptor |
| chr10_-_49701686 | 0.08 |
ENST00000417247.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chrX_-_1511617 | 0.07 |
ENST00000381401.5 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr11_+_72929402 | 0.07 |
ENST00000393596.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr19_-_43099070 | 0.06 |
ENST00000244336.5 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr5_-_179285848 | 0.06 |
ENST00000403396.2 ENST00000292586.6 |
C5orf45 |
chromosome 5 open reading frame 45 |
| chr4_+_72052964 | 0.05 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr19_+_33182823 | 0.04 |
ENST00000397061.3 |
NUDT19 |
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr1_-_216978709 | 0.04 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr5_-_179285785 | 0.04 |
ENST00000520698.1 ENST00000518235.1 ENST00000376931.2 ENST00000518219.1 ENST00000521333.1 ENST00000523084.1 |
C5orf45 |
chromosome 5 open reading frame 45 |
| chr1_+_66458072 | 0.04 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chrX_-_65858865 | 0.04 |
ENST00000374719.3 ENST00000450752.1 ENST00000451436.2 |
EDA2R |
ectodysplasin A2 receptor |
| chr3_-_196439065 | 0.03 |
ENST00000399942.4 ENST00000409690.3 |
CEP19 |
centrosomal protein 19kDa |
| chr21_+_19617140 | 0.03 |
ENST00000299295.2 ENST00000338326.3 |
CHODL |
chondrolectin |
| chr1_+_156124162 | 0.03 |
ENST00000368282.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr21_+_41239243 | 0.02 |
ENST00000328619.5 |
PCP4 |
Purkinje cell protein 4 |
| chr17_+_25936862 | 0.01 |
ENST00000582410.1 |
KSR1 |
kinase suppressor of ras 1 |
| chr20_+_306177 | 0.01 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr12_-_123215306 | 0.01 |
ENST00000356987.2 ENST00000436083.2 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
| chr11_-_62783276 | 0.00 |
ENST00000535878.1 ENST00000545207.1 |
SLC22A8 |
solute carrier family 22 (organic anion transporter), member 8 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.7 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 0.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 1.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 2.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.4 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.1 | 2.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.5 | 1.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 2.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 1.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 1.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.5 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 1.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.2 | GO:0044416 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 2.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 2.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 3.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.3 | 1.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |