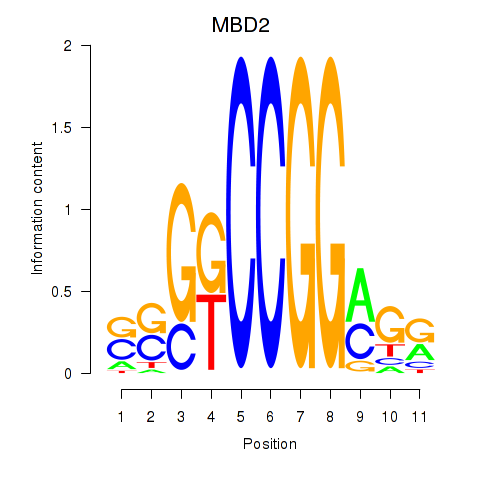
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MBD2
Z-value: 1.51
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | MBD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51751132_51751158 | -0.44 | 8.6e-02 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_9346892 | 4.47 |
ENST00000281419.3 ENST00000315273.4 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr1_+_99127225 | 4.12 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr2_+_173292390 | 4.03 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr1_+_99127265 | 4.02 |
ENST00000306121.3 |
SNX7 |
sorting nexin 7 |
| chr3_-_185542761 | 3.73 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_-_185542817 | 3.53 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_-_149688896 | 3.23 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr3_-_149688502 | 3.17 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr13_+_27131798 | 2.94 |
ENST00000361042.4 |
WASF3 |
WAS protein family, member 3 |
| chr10_-_17659234 | 2.90 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr13_+_27131887 | 2.80 |
ENST00000335327.5 |
WASF3 |
WAS protein family, member 3 |
| chr11_-_2160180 | 2.73 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr2_+_173292301 | 2.64 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr11_-_2160611 | 2.62 |
ENST00000416167.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr8_+_30241934 | 2.43 |
ENST00000538486.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr9_-_74383799 | 2.39 |
ENST00000377044.4 |
TMEM2 |
transmembrane protein 2 |
| chr11_+_69455855 | 2.22 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr13_+_98795434 | 2.19 |
ENST00000376586.2 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr13_+_98795505 | 2.17 |
ENST00000319562.6 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr10_-_17659357 | 2.10 |
ENST00000326961.6 ENST00000361271.3 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_+_30241995 | 2.06 |
ENST00000397323.4 ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS |
RNA binding protein with multiple splicing |
| chr12_-_124457257 | 2.01 |
ENST00000545891.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr13_-_80915059 | 1.94 |
ENST00000377104.3 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
| chr14_+_21538429 | 1.94 |
ENST00000298694.4 ENST00000555038.1 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr11_-_75236867 | 1.93 |
ENST00000376282.3 ENST00000336898.3 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr2_+_236402669 | 1.93 |
ENST00000409457.1 ENST00000336665.5 ENST00000304032.8 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr14_+_21538517 | 1.91 |
ENST00000298693.3 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr6_+_107811162 | 1.74 |
ENST00000317357.5 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
| chr12_+_19282643 | 1.71 |
ENST00000317589.4 ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr18_-_812517 | 1.69 |
ENST00000584307.1 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr16_-_22385901 | 1.64 |
ENST00000268383.2 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
| chr17_+_19552036 | 1.63 |
ENST00000581518.1 ENST00000395575.2 ENST00000584332.2 ENST00000339618.4 ENST00000579855.1 |
ALDH3A2 |
aldehyde dehydrogenase 3 family, member A2 |
| chr3_-_124774802 | 1.61 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr18_-_812231 | 1.58 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr19_+_18496957 | 1.56 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr12_-_124457371 | 1.54 |
ENST00000238156.3 ENST00000545037.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr10_+_104404218 | 1.51 |
ENST00000302424.7 |
TRIM8 |
tripartite motif containing 8 |
| chr3_+_33155525 | 1.51 |
ENST00000449224.1 |
CRTAP |
cartilage associated protein |
| chr6_+_31865552 | 1.50 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr19_-_19051103 | 1.45 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr17_-_27278304 | 1.43 |
ENST00000577226.1 |
PHF12 |
PHD finger protein 12 |
| chr10_-_79686284 | 1.43 |
ENST00000372391.2 ENST00000372388.2 |
DLG5 |
discs, large homolog 5 (Drosophila) |
| chr1_+_82266053 | 1.43 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr16_-_4166186 | 1.41 |
ENST00000294016.3 |
ADCY9 |
adenylate cyclase 9 |
| chr19_-_11373128 | 1.40 |
ENST00000294618.7 |
DOCK6 |
dedicator of cytokinesis 6 |
| chr17_-_62658186 | 1.39 |
ENST00000262435.9 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
| chr14_-_103523745 | 1.38 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr12_-_115121962 | 1.37 |
ENST00000349155.2 |
TBX3 |
T-box 3 |
| chr12_-_44200146 | 1.35 |
ENST00000395510.2 ENST00000325127.4 |
TWF1 |
twinfilin actin-binding protein 1 |
| chr10_+_129845823 | 1.34 |
ENST00000306042.5 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
| chr6_+_148663729 | 1.34 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr5_-_39425290 | 1.33 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_43232649 | 1.27 |
ENST00000372526.2 ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr19_-_36643329 | 1.24 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr21_-_27542972 | 1.23 |
ENST00000346798.3 ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP |
amyloid beta (A4) precursor protein |
| chr7_+_94285637 | 1.19 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr5_-_127873659 | 1.16 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr3_+_33155444 | 1.15 |
ENST00000320954.6 |
CRTAP |
cartilage associated protein |
| chr12_-_44200052 | 1.14 |
ENST00000548315.1 ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1 |
twinfilin actin-binding protein 1 |
| chr9_-_136933134 | 1.11 |
ENST00000303407.7 |
BRD3 |
bromodomain containing 3 |
| chr7_+_40174565 | 1.08 |
ENST00000309930.5 ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10 |
succinylCoA:glutarate-CoA transferase |
| chr19_+_36606654 | 1.07 |
ENST00000588385.1 ENST00000585746.1 |
TBCB |
tubulin folding cofactor B |
| chr20_-_47804894 | 1.05 |
ENST00000371828.3 ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
| chr7_-_94285402 | 1.03 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr11_-_115375107 | 1.03 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr5_-_176900610 | 1.03 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chr2_-_11484710 | 1.00 |
ENST00000315872.6 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
| chr17_+_48133459 | 0.98 |
ENST00000320031.8 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr10_+_128593978 | 0.98 |
ENST00000280333.6 |
DOCK1 |
dedicator of cytokinesis 1 |
| chr7_-_94285472 | 0.97 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr1_-_9884011 | 0.96 |
ENST00000361311.4 |
CLSTN1 |
calsyntenin 1 |
| chr19_+_36606354 | 0.95 |
ENST00000589996.1 ENST00000591296.1 |
TBCB |
tubulin folding cofactor B |
| chr2_+_241375069 | 0.95 |
ENST00000264039.2 |
GPC1 |
glypican 1 |
| chr2_+_110551851 | 0.94 |
ENST00000272454.6 ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5 |
RANBP2-like and GRIP domain containing 5 |
| chr18_-_51750948 | 0.94 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr7_-_94285511 | 0.92 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr12_+_120972147 | 0.90 |
ENST00000325954.4 ENST00000542438.1 |
RNF10 |
ring finger protein 10 |
| chr11_-_76381781 | 0.89 |
ENST00000260061.5 ENST00000404995.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr5_+_14143728 | 0.88 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr3_-_133969437 | 0.88 |
ENST00000460933.1 ENST00000296084.4 |
RYK |
receptor-like tyrosine kinase |
| chr19_+_56152262 | 0.86 |
ENST00000325333.5 ENST00000590190.1 |
ZNF580 |
zinc finger protein 580 |
| chr19_+_36606933 | 0.84 |
ENST00000586868.1 |
TBCB |
tubulin folding cofactor B |
| chr14_-_89883412 | 0.84 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr22_-_50746027 | 0.83 |
ENST00000425954.1 ENST00000449103.1 |
PLXNB2 |
plexin B2 |
| chr17_+_46126135 | 0.82 |
ENST00000361665.3 ENST00000585062.1 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr6_+_139456226 | 0.81 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr19_-_1568057 | 0.80 |
ENST00000402693.4 ENST00000388824.6 |
MEX3D |
mex-3 RNA binding family member D |
| chrX_-_134049262 | 0.80 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr8_-_119124045 | 0.78 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr18_+_56530794 | 0.78 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr14_+_102829300 | 0.77 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr12_+_19282713 | 0.76 |
ENST00000299275.6 ENST00000539256.1 ENST00000538714.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr8_+_37654693 | 0.75 |
ENST00000412232.2 |
GPR124 |
G protein-coupled receptor 124 |
| chr9_-_130617029 | 0.74 |
ENST00000373203.4 |
ENG |
endoglin |
| chr22_-_50746001 | 0.73 |
ENST00000359337.4 |
PLXNB2 |
plexin B2 |
| chr11_-_89224508 | 0.73 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr17_+_46125707 | 0.73 |
ENST00000584137.1 ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr12_-_48398104 | 0.72 |
ENST00000337299.6 ENST00000380518.3 |
COL2A1 |
collagen, type II, alpha 1 |
| chr11_-_89224488 | 0.71 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr20_-_56284816 | 0.71 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr17_-_80231557 | 0.71 |
ENST00000392334.2 ENST00000314028.6 |
CSNK1D |
casein kinase 1, delta |
| chr3_-_133969673 | 0.70 |
ENST00000427044.2 |
RYK |
receptor-like tyrosine kinase |
| chr17_-_41623075 | 0.70 |
ENST00000545089.1 |
ETV4 |
ets variant 4 |
| chr3_-_38691119 | 0.70 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr3_-_98620500 | 0.69 |
ENST00000326840.6 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
| chr11_-_89224299 | 0.67 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr10_+_75545391 | 0.67 |
ENST00000604524.1 ENST00000605216.1 ENST00000398706.2 |
ZSWIM8 |
zinc finger, SWIM-type containing 8 |
| chr7_-_558876 | 0.65 |
ENST00000354513.5 ENST00000402802.3 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
| chr1_+_19923454 | 0.65 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr17_+_68165657 | 0.65 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_62369291 | 0.65 |
ENST00000278823.2 |
MTA2 |
metastasis associated 1 family, member 2 |
| chr2_+_54198210 | 0.64 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr3_-_52001448 | 0.63 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr1_+_84543734 | 0.63 |
ENST00000370689.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_-_130616915 | 0.62 |
ENST00000344849.3 |
ENG |
endoglin |
| chr15_-_73925651 | 0.61 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr11_-_89224638 | 0.61 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr11_+_43380459 | 0.60 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chrX_+_9983602 | 0.60 |
ENST00000380861.4 |
WWC3 |
WWC family member 3 |
| chr17_-_41623259 | 0.58 |
ENST00000538265.1 ENST00000591713.1 |
ETV4 |
ets variant 4 |
| chr10_+_75545329 | 0.58 |
ENST00000604729.1 ENST00000603114.1 |
ZSWIM8 |
zinc finger, SWIM-type containing 8 |
| chr14_+_74416989 | 0.57 |
ENST00000334571.2 ENST00000554920.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr17_-_42277203 | 0.57 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr1_+_156698234 | 0.57 |
ENST00000368218.4 ENST00000368216.4 |
RRNAD1 |
ribosomal RNA adenine dimethylase domain containing 1 |
| chr12_-_57824739 | 0.57 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chr8_-_145688231 | 0.56 |
ENST00000530374.1 |
CYHR1 |
cysteine/histidine-rich 1 |
| chr1_+_43855560 | 0.56 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chrX_+_152953505 | 0.55 |
ENST00000253122.5 |
SLC6A8 |
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr17_-_80231300 | 0.53 |
ENST00000398519.5 ENST00000580446.1 |
CSNK1D |
casein kinase 1, delta |
| chr3_+_197518100 | 0.53 |
ENST00000438796.2 ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr2_+_5832799 | 0.52 |
ENST00000322002.3 |
SOX11 |
SRY (sex determining region Y)-box 11 |
| chr11_-_89224139 | 0.51 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr2_-_178257401 | 0.51 |
ENST00000464747.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr6_+_1610681 | 0.50 |
ENST00000380874.2 |
FOXC1 |
forkhead box C1 |
| chr9_+_103235365 | 0.50 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr3_+_142442841 | 0.49 |
ENST00000476941.1 ENST00000273482.6 |
TRPC1 |
transient receptor potential cation channel, subfamily C, member 1 |
| chr18_-_72265035 | 0.49 |
ENST00000585279.1 ENST00000580048.1 |
LINC00909 |
long intergenic non-protein coding RNA 909 |
| chr6_-_91296737 | 0.48 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr11_-_72353451 | 0.47 |
ENST00000376450.3 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr2_+_219264466 | 0.47 |
ENST00000273062.2 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr14_-_23564320 | 0.46 |
ENST00000605057.1 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
| chr1_-_156252590 | 0.46 |
ENST00000361813.5 ENST00000368267.5 |
SMG5 |
SMG5 nonsense mediated mRNA decay factor |
| chr17_-_41623009 | 0.46 |
ENST00000393664.2 |
ETV4 |
ets variant 4 |
| chr6_-_91296602 | 0.45 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr7_+_86781847 | 0.45 |
ENST00000432366.2 ENST00000423590.2 ENST00000394703.5 |
DMTF1 |
cyclin D binding myb-like transcription factor 1 |
| chr1_+_156338993 | 0.45 |
ENST00000368249.1 ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG |
Rh family, B glycoprotein (gene/pseudogene) |
| chr1_+_6845384 | 0.45 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr20_-_48532019 | 0.44 |
ENST00000289431.5 |
SPATA2 |
spermatogenesis associated 2 |
| chr3_-_88108212 | 0.44 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr2_-_220408430 | 0.43 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr7_+_86781677 | 0.42 |
ENST00000331242.7 ENST00000394702.3 ENST00000413276.2 ENST00000446796.2 ENST00000411766.2 ENST00000420131.1 ENST00000414630.2 ENST00000453049.1 ENST00000428819.1 ENST00000448598.1 ENST00000449088.3 ENST00000430405.3 |
DMTF1 |
cyclin D binding myb-like transcription factor 1 |
| chrX_+_24167828 | 0.41 |
ENST00000379188.3 ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX |
zinc finger protein, X-linked |
| chr16_-_49890016 | 0.40 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr6_-_10415470 | 0.38 |
ENST00000379604.2 ENST00000379613.3 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr20_+_34894247 | 0.38 |
ENST00000373913.3 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr22_-_20004330 | 0.38 |
ENST00000263207.3 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr16_-_89787360 | 0.38 |
ENST00000389386.3 |
VPS9D1 |
VPS9 domain containing 1 |
| chrX_-_40594755 | 0.38 |
ENST00000324817.1 |
MED14 |
mediator complex subunit 14 |
| chr1_-_23857698 | 0.38 |
ENST00000361729.2 |
E2F2 |
E2F transcription factor 2 |
| chr14_-_77787198 | 0.37 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr7_-_139876812 | 0.36 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr2_-_9695847 | 0.36 |
ENST00000310823.3 ENST00000497134.1 |
ADAM17 |
ADAM metallopeptidase domain 17 |
| chr6_+_29795595 | 0.35 |
ENST00000360323.6 ENST00000376818.3 ENST00000376815.3 |
HLA-G |
major histocompatibility complex, class I, G |
| chr19_-_16653325 | 0.34 |
ENST00000546361.2 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr8_-_93115445 | 0.34 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr11_+_64851729 | 0.34 |
ENST00000526791.1 ENST00000526945.1 |
ZFPL1 |
zinc finger protein-like 1 |
| chr5_-_72744336 | 0.34 |
ENST00000499003.3 |
FOXD1 |
forkhead box D1 |
| chr1_-_21503337 | 0.34 |
ENST00000400422.1 ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr12_+_111051832 | 0.34 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr12_+_120972606 | 0.33 |
ENST00000413266.2 |
RNF10 |
ring finger protein 10 |
| chr19_-_16653226 | 0.32 |
ENST00000198939.6 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr12_+_111051902 | 0.31 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr14_-_93651186 | 0.31 |
ENST00000556883.1 ENST00000298894.4 |
MOAP1 |
modulator of apoptosis 1 |
| chr2_-_118771701 | 0.31 |
ENST00000376300.2 ENST00000319432.5 |
CCDC93 |
coiled-coil domain containing 93 |
| chr5_-_180076613 | 0.30 |
ENST00000261937.6 ENST00000393347.3 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr4_-_11430221 | 0.30 |
ENST00000514690.1 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr14_-_105262016 | 0.30 |
ENST00000407796.2 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr8_-_101734308 | 0.30 |
ENST00000519004.1 ENST00000519363.1 ENST00000520142.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr9_+_137967366 | 0.30 |
ENST00000252854.4 |
OLFM1 |
olfactomedin 1 |
| chrX_+_68725084 | 0.29 |
ENST00000252338.4 |
FAM155B |
family with sequence similarity 155, member B |
| chr14_-_105262055 | 0.29 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr20_-_48532046 | 0.29 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr17_+_43971643 | 0.29 |
ENST00000344290.5 ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT |
microtubule-associated protein tau |
| chr19_+_50353944 | 0.29 |
ENST00000594151.1 ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr6_-_10415218 | 0.28 |
ENST00000466073.1 ENST00000498450.1 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr12_+_122516626 | 0.28 |
ENST00000319080.7 |
MLXIP |
MLX interacting protein |
| chr19_-_2151523 | 0.28 |
ENST00000350812.6 ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1 |
adaptor-related protein complex 3, delta 1 subunit |
| chr5_-_180076580 | 0.28 |
ENST00000502649.1 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr11_+_76092353 | 0.28 |
ENST00000530460.1 ENST00000321844.4 |
RP11-111M22.2 |
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr7_-_19157248 | 0.27 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr14_+_103851712 | 0.27 |
ENST00000440884.3 ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
| chr19_+_50354393 | 0.27 |
ENST00000391842.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr1_+_36023035 | 0.26 |
ENST00000373253.3 |
NCDN |
neurochondrin |
| chr11_+_66025938 | 0.26 |
ENST00000394066.2 |
KLC2 |
kinesin light chain 2 |
| chr11_-_62414070 | 0.26 |
ENST00000540933.1 ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB |
glucosidase, alpha; neutral AB |
| chr11_+_111473108 | 0.25 |
ENST00000304987.3 |
SIK2 |
salt-inducible kinase 2 |
| chr9_-_112260531 | 0.25 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr9_-_131644202 | 0.25 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr1_-_36022979 | 0.25 |
ENST00000469892.1 ENST00000325722.3 |
KIAA0319L |
KIAA0319-like |
| chr3_+_50273625 | 0.25 |
ENST00000536647.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.6 | 1.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.5 | 1.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.5 | 6.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 1.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 3.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.3 | 1.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 2.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.7 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 2.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 7.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.8 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 8.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 7.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 1.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.9 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 2.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 5.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 3.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 2.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.6 | 1.9 | GO:0060437 | lung growth(GO:0060437) |
| 0.6 | 5.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.5 | 6.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 2.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.5 | 6.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) nail development(GO:0035878) |
| 0.5 | 1.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.5 | 1.4 | GO:0060926 | cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.4 | 1.6 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.4 | 1.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 2.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 1.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.3 | 1.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 0.7 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.3 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 2.8 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.3 | 1.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 4.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 1.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 6.9 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 1.6 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.2 | 1.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 0.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 0.5 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 0.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 1.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.5 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 1.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.4 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 3.3 | GO:0036119 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.3 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.6 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 1.2 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 3.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 1.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 2.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.4 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.7 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 1.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 4.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.2 | GO:0072092 | olfactory bulb mitral cell layer development(GO:0061034) ureteric bud invasion(GO:0072092) |
| 0.1 | 7.4 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.1 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 1.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.7 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0002767 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 1.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.6 | GO:0090037 | regulation of blood vessel remodeling(GO:0060312) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 1.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.3 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.9 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.6 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.7 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 3.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 7.7 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 7.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 4.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 7.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 3.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 6.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.5 | 1.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.4 | 2.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.0 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.2 | 3.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 4.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 5.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 6.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 6.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 4.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 10.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 5.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.2 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |