Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MECOM
Z-value: 0.72
Transcription factors associated with MECOM
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MECOM
|
ENSG00000085276.13 | MECOM |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MECOM | hg19_v2_chr3_-_168864427_168864464, hg19_v2_chr3_-_169381183_169381258, hg19_v2_chr3_-_168865522_168865536 | 0.09 | 7.3e-01 | Click! |
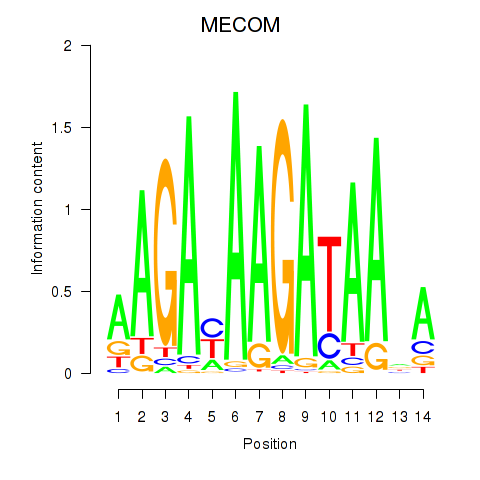
Activity profile of MECOM motif
Sorted Z-values of MECOM motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MECOM
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74275057 | 2.08 |
ENST00000511370.1 |
ALB |
albumin |
| chr3_-_145968923 | 1.91 |
ENST00000493382.1 ENST00000354952.2 ENST00000383083.2 |
PLSCR4 |
phospholipid scramblase 4 |
| chr8_-_6420777 | 1.74 |
ENST00000415216.1 |
ANGPT2 |
angiopoietin 2 |
| chr3_-_145968857 | 1.73 |
ENST00000433593.2 ENST00000476202.1 ENST00000460885.1 |
PLSCR4 |
phospholipid scramblase 4 |
| chr17_+_53344945 | 1.70 |
ENST00000575345.1 |
HLF |
hepatic leukemia factor |
| chr1_-_161193349 | 1.65 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr5_+_92919043 | 1.60 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr5_+_102201722 | 1.29 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_102201509 | 1.20 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr12_-_15038779 | 1.18 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr7_+_17338239 | 1.18 |
ENST00000242057.4 |
AHR |
aryl hydrocarbon receptor |
| chr8_-_6420930 | 1.15 |
ENST00000325203.5 |
ANGPT2 |
angiopoietin 2 |
| chr4_+_74301880 | 1.13 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr6_-_52860171 | 1.11 |
ENST00000370963.4 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr3_-_149093499 | 1.08 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr8_-_6420565 | 1.06 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr2_-_56150910 | 1.00 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr3_+_148583043 | 0.99 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr8_-_6420759 | 0.91 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr4_+_155484155 | 0.85 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr1_-_146696901 | 0.82 |
ENST00000369272.3 ENST00000441068.2 |
FMO5 |
flavin containing monooxygenase 5 |
| chr5_+_102201430 | 0.81 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_15525426 | 0.69 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr1_+_82266053 | 0.68 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr9_-_13175823 | 0.67 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chrX_-_106243451 | 0.64 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr2_+_242289502 | 0.62 |
ENST00000451310.1 |
SEPT2 |
septin 2 |
| chr4_+_155484103 | 0.61 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr6_-_52859968 | 0.56 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr13_+_76334567 | 0.54 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr8_-_93029865 | 0.53 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr9_-_107690420 | 0.52 |
ENST00000423487.2 ENST00000374733.1 ENST00000374736.3 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chrX_-_43741594 | 0.51 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr11_+_17316870 | 0.46 |
ENST00000458064.2 |
NUCB2 |
nucleobindin 2 |
| chr2_+_37571717 | 0.45 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chrX_-_10588459 | 0.45 |
ENST00000380782.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr1_+_171060018 | 0.44 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr13_+_102142296 | 0.42 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_+_153019069 | 0.42 |
ENST00000532295.1 |
MYCT1 |
myc target 1 |
| chr14_+_22458631 | 0.42 |
ENST00000390444.1 |
TRAV16 |
T cell receptor alpha variable 16 |
| chr3_-_178984759 | 0.41 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr6_+_153019023 | 0.41 |
ENST00000367245.5 ENST00000529453.1 |
MYCT1 |
myc target 1 |
| chr7_-_100844193 | 0.41 |
ENST00000440203.2 ENST00000379423.3 ENST00000223114.4 |
MOGAT3 |
monoacylglycerol O-acyltransferase 3 |
| chr19_-_47735918 | 0.40 |
ENST00000449228.1 ENST00000300880.7 ENST00000341983.4 |
BBC3 |
BCL2 binding component 3 |
| chrX_+_67718863 | 0.39 |
ENST00000374622.2 |
YIPF6 |
Yip1 domain family, member 6 |
| chrX_-_10588595 | 0.39 |
ENST00000423614.1 ENST00000317552.4 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr20_+_36012051 | 0.38 |
ENST00000373567.2 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr14_-_25479811 | 0.37 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr11_+_22696314 | 0.36 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr16_-_73093597 | 0.36 |
ENST00000397992.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr1_-_98386543 | 0.36 |
ENST00000423006.2 ENST00000370192.3 ENST00000306031.5 |
DPYD |
dihydropyrimidine dehydrogenase |
| chr14_+_64680854 | 0.36 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr4_-_40859132 | 0.34 |
ENST00000543538.1 ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr3_+_99357319 | 0.33 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr17_-_56494882 | 0.32 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chrX_+_66764375 | 0.30 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr17_-_56494908 | 0.30 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr9_+_134378289 | 0.29 |
ENST00000423007.1 ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1 |
protein-O-mannosyltransferase 1 |
| chr17_-_56494713 | 0.28 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chrX_+_102883620 | 0.28 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr12_+_57522258 | 0.28 |
ENST00000553277.1 ENST00000243077.3 |
LRP1 |
low density lipoprotein receptor-related protein 1 |
| chr2_+_172544294 | 0.27 |
ENST00000358002.6 ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_172543967 | 0.27 |
ENST00000534253.2 ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_172544011 | 0.27 |
ENST00000508530.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_-_1629176 | 0.26 |
ENST00000366424.2 |
AC144450.2 |
AC144450.2 |
| chr2_+_189839046 | 0.26 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr2_+_172544182 | 0.25 |
ENST00000409197.1 ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_-_48894762 | 0.25 |
ENST00000600980.1 ENST00000330720.2 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr14_+_24584508 | 0.25 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chrX_+_2959512 | 0.24 |
ENST00000381127.1 |
ARSF |
arylsulfatase F |
| chr14_-_24584138 | 0.24 |
ENST00000558280.1 ENST00000561028.1 |
NRL |
neural retina leucine zipper |
| chr5_+_140739537 | 0.24 |
ENST00000522605.1 |
PCDHGB2 |
protocadherin gamma subfamily B, 2 |
| chr11_-_18656028 | 0.24 |
ENST00000336349.5 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr1_-_208084729 | 0.24 |
ENST00000310833.7 ENST00000356522.4 |
CD34 |
CD34 molecule |
| chr22_+_29138013 | 0.23 |
ENST00000216027.3 ENST00000398941.2 |
HSCB |
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chrX_+_86772787 | 0.21 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr20_-_45985464 | 0.20 |
ENST00000458360.2 ENST00000262975.4 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr2_+_55746746 | 0.20 |
ENST00000406691.3 ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104 |
coiled-coil domain containing 104 |
| chr20_-_45984401 | 0.20 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr2_+_172543919 | 0.18 |
ENST00000452242.1 ENST00000340296.4 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_-_71758602 | 0.17 |
ENST00000568954.1 |
PHLPP2 |
PH domain and leucine rich repeat protein phosphatase 2 |
| chr3_-_45883558 | 0.17 |
ENST00000445698.1 ENST00000296135.6 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr7_-_115670804 | 0.17 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr12_+_72080253 | 0.16 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chr6_+_31515337 | 0.15 |
ENST00000376148.4 ENST00000376145.4 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr3_-_27498235 | 0.15 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr20_-_45985414 | 0.15 |
ENST00000461685.1 ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr9_+_82186682 | 0.15 |
ENST00000376552.2 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr20_-_45985172 | 0.14 |
ENST00000536340.1 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr16_-_3930724 | 0.14 |
ENST00000262367.5 |
CREBBP |
CREB binding protein |
| chr2_-_231989808 | 0.14 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr19_-_46366392 | 0.14 |
ENST00000598059.1 ENST00000594293.1 ENST00000245934.7 |
SYMPK |
symplekin |
| chr5_-_115910091 | 0.13 |
ENST00000257414.8 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr16_-_50402836 | 0.13 |
ENST00000394688.3 |
BRD7 |
bromodomain containing 7 |
| chr13_-_46626847 | 0.12 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr10_-_29923893 | 0.11 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr10_+_57358750 | 0.11 |
ENST00000512524.2 |
MTRNR2L5 |
MT-RNR2-like 5 |
| chr12_-_81763127 | 0.11 |
ENST00000541017.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr8_-_133637624 | 0.11 |
ENST00000522789.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr2_+_97779233 | 0.11 |
ENST00000461153.2 ENST00000420699.2 |
ANKRD36 |
ankyrin repeat domain 36 |
| chr5_+_74807886 | 0.10 |
ENST00000514296.1 |
POLK |
polymerase (DNA directed) kappa |
| chr6_+_29624898 | 0.09 |
ENST00000396704.3 ENST00000483013.1 ENST00000490427.1 ENST00000416766.2 ENST00000376891.4 ENST00000376898.3 ENST00000396701.2 ENST00000494692.1 ENST00000431798.2 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr2_+_108145913 | 0.09 |
ENST00000443205.1 |
AC096669.3 |
AC096669.3 |
| chr3_+_39448180 | 0.09 |
ENST00000301821.6 ENST00000458478.1 ENST00000443003.1 |
RPSA |
ribosomal protein SA |
| chr7_+_97361388 | 0.09 |
ENST00000350485.4 ENST00000346867.4 |
TAC1 |
tachykinin, precursor 1 |
| chr11_+_61008514 | 0.09 |
ENST00000312403.5 |
PGA5 |
pepsinogen 5, group I (pepsinogen A) |
| chr19_+_36630454 | 0.09 |
ENST00000246533.3 |
CAPNS1 |
calpain, small subunit 1 |
| chr15_-_48470558 | 0.08 |
ENST00000324324.7 |
MYEF2 |
myelin expression factor 2 |
| chrX_-_85302531 | 0.08 |
ENST00000537751.1 ENST00000358786.4 ENST00000357749.2 |
CHM |
choroideremia (Rab escort protein 1) |
| chr16_-_50402690 | 0.08 |
ENST00000394689.2 |
BRD7 |
bromodomain containing 7 |
| chr5_+_54320078 | 0.08 |
ENST00000231009.2 |
GZMK |
granzyme K (granzyme 3; tryptase II) |
| chr12_+_15699286 | 0.08 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr17_-_37308824 | 0.08 |
ENST00000415163.1 ENST00000441877.1 ENST00000444911.2 |
PLXDC1 |
plexin domain containing 1 |
| chr8_-_39695719 | 0.08 |
ENST00000347580.4 ENST00000379853.2 ENST00000521880.1 |
ADAM2 |
ADAM metallopeptidase domain 2 |
| chr9_+_82186872 | 0.08 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_-_102701740 | 0.08 |
ENST00000561150.1 ENST00000522867.1 |
MOK |
MOK protein kinase |
| chr11_+_60989688 | 0.08 |
ENST00000378149.3 ENST00000422676.2 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr12_+_72079842 | 0.08 |
ENST00000266673.5 ENST00000550524.1 |
TMEM19 |
transmembrane protein 19 |
| chr3_-_38992052 | 0.08 |
ENST00000302328.3 ENST00000450244.1 ENST00000444237.2 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
| chr16_+_10837643 | 0.08 |
ENST00000574334.1 ENST00000283027.5 ENST00000433392.2 |
NUBP1 |
nucleotide binding protein 1 |
| chr8_-_39695764 | 0.07 |
ENST00000265708.4 |
ADAM2 |
ADAM metallopeptidase domain 2 |
| chr12_+_96588279 | 0.07 |
ENST00000552142.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr15_+_71185148 | 0.07 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr1_+_15802594 | 0.07 |
ENST00000375910.3 |
CELA2B |
chymotrypsin-like elastase family, member 2B |
| chr12_+_21207503 | 0.07 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_-_81763184 | 0.06 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_172422026 | 0.06 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_+_160160346 | 0.06 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chrX_+_155227246 | 0.05 |
ENST00000244174.5 ENST00000424344.3 |
IL9R |
interleukin 9 receptor |
| chr16_-_90096309 | 0.05 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr6_-_10882174 | 0.05 |
ENST00000379491.4 |
GCM2 |
glial cells missing homolog 2 (Drosophila) |
| chr20_+_36932521 | 0.04 |
ENST00000262865.4 |
BPI |
bactericidal/permeability-increasing protein |
| chr2_+_27435734 | 0.04 |
ENST00000419744.1 |
ATRAID |
all-trans retinoic acid-induced differentiation factor |
| chr6_-_154831779 | 0.04 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr16_+_9449445 | 0.04 |
ENST00000564305.1 |
RP11-243A14.1 |
RP11-243A14.1 |
| chr9_+_33240157 | 0.04 |
ENST00000379721.3 |
SPINK4 |
serine peptidase inhibitor, Kazal type 4 |
| chr4_-_74847800 | 0.04 |
ENST00000296029.3 |
PF4 |
platelet factor 4 |
| chr6_-_31514516 | 0.04 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr13_-_113242439 | 0.04 |
ENST00000375669.3 ENST00000261965.3 |
TUBGCP3 |
tubulin, gamma complex associated protein 3 |
| chr3_+_147127142 | 0.04 |
ENST00000282928.4 |
ZIC1 |
Zic family member 1 |
| chr12_+_21168630 | 0.04 |
ENST00000421593.2 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_184971853 | 0.03 |
ENST00000231887.3 |
EHHADH |
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr7_-_15601595 | 0.03 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr22_+_36113919 | 0.03 |
ENST00000249044.2 |
APOL5 |
apolipoprotein L, 5 |
| chr13_+_49280951 | 0.03 |
ENST00000282018.3 |
CYSLTR2 |
cysteinyl leukotriene receptor 2 |
| chr22_-_31688431 | 0.03 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr14_-_24551195 | 0.03 |
ENST00000560550.1 |
NRL |
neural retina leucine zipper |
| chr19_+_48497962 | 0.02 |
ENST00000596043.1 ENST00000597519.1 |
ELSPBP1 |
epididymal sperm binding protein 1 |
| chr4_-_77997126 | 0.02 |
ENST00000537948.1 ENST00000507788.1 ENST00000237654.4 |
CCNI |
cyclin I |
| chr4_-_88450244 | 0.02 |
ENST00000503414.1 |
SPARCL1 |
SPARC-like 1 (hevin) |
| chr8_+_134203303 | 0.02 |
ENST00000519433.1 ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr19_-_10614386 | 0.02 |
ENST00000171111.5 |
KEAP1 |
kelch-like ECH-associated protein 1 |
| chr17_-_43138357 | 0.02 |
ENST00000342350.5 |
DCAKD |
dephospho-CoA kinase domain containing |
| chr19_-_9785743 | 0.01 |
ENST00000537617.1 ENST00000589542.1 ENST00000590155.1 ENST00000541032.1 ENST00000588653.1 ENST00000448622.1 ENST00000453792.2 |
ZNF562 |
zinc finger protein 562 |
| chr1_-_92949505 | 0.01 |
ENST00000370332.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr17_-_10276319 | 0.01 |
ENST00000252172.4 ENST00000418404.3 |
MYH13 |
myosin, heavy chain 13, skeletal muscle |
| chr14_-_75179774 | 0.01 |
ENST00000555249.1 ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1 AC007956.1 |
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr1_+_25664408 | 0.01 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr5_-_147162078 | 0.01 |
ENST00000507386.1 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
| chr6_+_31514622 | 0.01 |
ENST00000376146.4 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr19_+_47538560 | 0.00 |
ENST00000439365.2 ENST00000594670.1 |
NPAS1 |
neuronal PAS domain protein 1 |
| chr8_+_134203273 | 0.00 |
ENST00000250160.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr5_+_140165876 | 0.00 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr3_+_121311966 | 0.00 |
ENST00000338040.4 |
FBXO40 |
F-box protein 40 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.4 | 1.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 3.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.8 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.2 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 2.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 4.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.8 | 3.3 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.7 | 2.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.5 | 1.6 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.3 | 3.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 0.9 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 1.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.7 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 1.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.3 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.0 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:0038001 | paracrine signaling(GO:0038001) glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 0.3 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 1.6 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 1.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 1.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 3.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |