Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
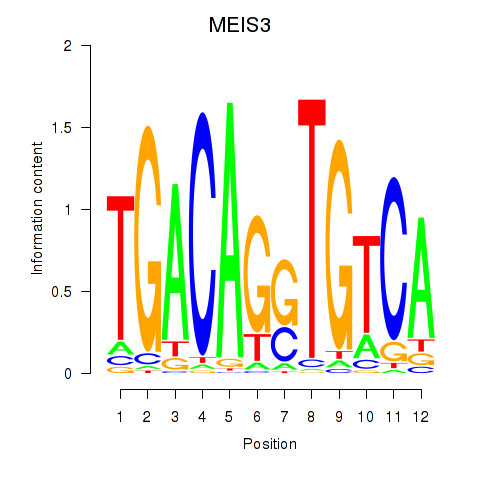
Results for MEIS3_TGIF2LX
Z-value: 0.70
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | MEIS3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGIF2LX |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_8775466 | 1.35 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr11_-_2170786 | 1.27 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr6_-_52860171 | 1.13 |
ENST00000370963.4 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr2_+_24714729 | 0.96 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr16_+_30212378 | 0.96 |
ENST00000569485.1 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr6_-_52859968 | 0.75 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr10_-_105212141 | 0.73 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr10_-_105212059 | 0.72 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr3_-_49726486 | 0.68 |
ENST00000449682.2 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr18_-_52989525 | 0.65 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr11_-_76381029 | 0.64 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr17_+_41003166 | 0.61 |
ENST00000308423.2 |
AOC3 |
amine oxidase, copper containing 3 |
| chr1_-_153517473 | 0.57 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr6_-_112194484 | 0.56 |
ENST00000518295.1 ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr1_+_161736072 | 0.54 |
ENST00000367942.3 |
ATF6 |
activating transcription factor 6 |
| chr13_-_67802549 | 0.51 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr11_-_119187826 | 0.48 |
ENST00000264036.4 |
MCAM |
melanoma cell adhesion molecule |
| chr21_+_30502806 | 0.43 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr16_+_72090053 | 0.40 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr19_-_45826125 | 0.40 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr9_+_34990219 | 0.40 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr11_-_107590383 | 0.40 |
ENST00000525934.1 ENST00000531293.1 |
SLN |
sarcolipin |
| chr12_+_121647962 | 0.39 |
ENST00000542067.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr6_+_24495067 | 0.39 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr2_+_97202480 | 0.37 |
ENST00000357485.3 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr12_+_121647868 | 0.37 |
ENST00000359949.7 ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr6_+_151662815 | 0.35 |
ENST00000359755.5 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr3_-_3221358 | 0.33 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr5_+_155753745 | 0.31 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr2_+_97203082 | 0.30 |
ENST00000454558.2 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr5_+_15500280 | 0.29 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr19_+_35773242 | 0.28 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr11_-_72070206 | 0.25 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr16_+_29789561 | 0.25 |
ENST00000400752.4 |
ZG16 |
zymogen granule protein 16 |
| chr16_-_18468926 | 0.22 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr9_+_136501478 | 0.22 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr19_-_41256207 | 0.22 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr7_-_29234802 | 0.21 |
ENST00000449801.1 ENST00000409850.1 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr14_-_80677613 | 0.21 |
ENST00000556811.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr17_-_1395954 | 0.21 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chrX_+_153775821 | 0.21 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr12_+_104458235 | 0.21 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr19_+_12273866 | 0.21 |
ENST00000425827.1 ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136 |
zinc finger protein 136 |
| chr4_+_671711 | 0.21 |
ENST00000400159.2 |
MYL5 |
myosin, light chain 5, regulatory |
| chr11_-_76381781 | 0.20 |
ENST00000260061.5 ENST00000404995.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr20_+_306177 | 0.20 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr15_+_68924327 | 0.20 |
ENST00000543950.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chrX_-_11445856 | 0.20 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr10_+_104503727 | 0.19 |
ENST00000448841.1 |
WBP1L |
WW domain binding protein 1-like |
| chr11_+_61447845 | 0.19 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr2_-_198299726 | 0.19 |
ENST00000409915.4 ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1 |
splicing factor 3b, subunit 1, 155kDa |
| chr21_-_43735446 | 0.19 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr6_+_160769399 | 0.19 |
ENST00000392145.1 |
SLC22A3 |
solute carrier family 22 (organic cation transporter), member 3 |
| chr1_-_84326653 | 0.18 |
ENST00000413975.1 ENST00000417975.1 ENST00000419733.1 |
RP11-475O6.1 |
RP11-475O6.1 |
| chr19_-_4559814 | 0.17 |
ENST00000586582.1 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr12_+_100867486 | 0.17 |
ENST00000548884.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr22_+_44319619 | 0.17 |
ENST00000216180.3 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr4_+_128554081 | 0.17 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr17_+_48172639 | 0.16 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr6_+_160769300 | 0.15 |
ENST00000275300.2 |
SLC22A3 |
solute carrier family 22 (organic cation transporter), member 3 |
| chr11_+_1942580 | 0.15 |
ENST00000381558.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr12_+_100867694 | 0.15 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr10_-_7661623 | 0.15 |
ENST00000298441.6 |
ITIH5 |
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr19_-_58609570 | 0.14 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr11_+_67776012 | 0.13 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr22_-_31885514 | 0.13 |
ENST00000397525.1 |
EIF4ENIF1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr8_-_61880248 | 0.13 |
ENST00000525556.1 |
AC022182.3 |
AC022182.3 |
| chr20_+_306221 | 0.12 |
ENST00000342665.2 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chrX_+_17755563 | 0.12 |
ENST00000380045.3 ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1 |
sex comb on midleg-like 1 (Drosophila) |
| chr6_-_32557610 | 0.12 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr19_+_5455421 | 0.12 |
ENST00000222033.4 |
ZNRF4 |
zinc and ring finger 4 |
| chr1_+_79115503 | 0.12 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr2_-_55920952 | 0.12 |
ENST00000447944.2 |
PNPT1 |
polyribonucleotide nucleotidyltransferase 1 |
| chr1_+_100598691 | 0.11 |
ENST00000370143.1 ENST00000370141.2 |
TRMT13 |
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr17_-_33446735 | 0.11 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr5_-_131329918 | 0.11 |
ENST00000357096.1 ENST00000431707.1 ENST00000434099.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr17_-_33446820 | 0.11 |
ENST00000592577.1 ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D |
RAD51 paralog D |
| chr22_+_44319648 | 0.11 |
ENST00000423180.2 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr22_-_43355858 | 0.11 |
ENST00000402229.1 ENST00000407585.1 ENST00000453079.1 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_+_128634589 | 0.11 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_+_104535994 | 0.11 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr7_-_72936531 | 0.11 |
ENST00000339594.4 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr14_-_80677815 | 0.11 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr3_+_52719936 | 0.10 |
ENST00000418458.1 ENST00000394799.2 |
GNL3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_-_242089677 | 0.10 |
ENST00000405260.1 |
PASK |
PAS domain containing serine/threonine kinase |
| chr7_+_76139833 | 0.10 |
ENST00000257632.5 |
UPK3B |
uroplakin 3B |
| chr10_+_51187938 | 0.10 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chr21_+_34619079 | 0.10 |
ENST00000433395.2 |
AP000295.9 |
AP000295.9 |
| chr2_+_90139056 | 0.10 |
ENST00000492446.1 |
IGKV1D-16 |
immunoglobulin kappa variable 1D-16 |
| chr12_-_56615693 | 0.10 |
ENST00000394013.2 ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41 |
ring finger protein 41 |
| chr1_-_169680745 | 0.10 |
ENST00000236147.4 |
SELL |
selectin L |
| chr2_+_90248739 | 0.09 |
ENST00000468879.1 |
IGKV1D-43 |
immunoglobulin kappa variable 1D-43 |
| chr2_-_24149977 | 0.09 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr19_+_17982747 | 0.08 |
ENST00000222248.3 |
SLC5A5 |
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr11_+_63304273 | 0.08 |
ENST00000439013.2 ENST00000255688.3 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
| chr6_-_30128657 | 0.08 |
ENST00000449742.2 ENST00000376704.3 |
TRIM10 |
tripartite motif containing 10 |
| chr2_-_65593784 | 0.08 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr7_+_76139925 | 0.08 |
ENST00000394849.1 |
UPK3B |
uroplakin 3B |
| chr8_-_98290087 | 0.08 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr17_+_34431212 | 0.08 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr2_+_162087577 | 0.08 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr8_+_56014949 | 0.08 |
ENST00000327381.6 |
XKR4 |
XK, Kell blood group complex subunit-related family, member 4 |
| chr5_-_131330272 | 0.08 |
ENST00000379240.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr6_+_24495185 | 0.07 |
ENST00000348925.2 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr3_+_29322437 | 0.07 |
ENST00000434693.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr11_-_65325664 | 0.07 |
ENST00000301873.5 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr16_-_2004683 | 0.06 |
ENST00000268661.7 |
RPL3L |
ribosomal protein L3-like |
| chr4_+_175839506 | 0.06 |
ENST00000505141.1 ENST00000359240.3 ENST00000445694.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chrX_+_148622138 | 0.06 |
ENST00000450602.2 ENST00000441248.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr16_+_1306060 | 0.06 |
ENST00000397534.2 |
TPSD1 |
tryptase delta 1 |
| chr16_+_1306093 | 0.06 |
ENST00000211076.3 |
TPSD1 |
tryptase delta 1 |
| chr11_-_134123142 | 0.06 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr4_+_175839551 | 0.06 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr2_+_10560147 | 0.06 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr6_+_42883727 | 0.06 |
ENST00000304672.1 ENST00000441198.1 ENST00000446507.1 |
PTCRA |
pre T-cell antigen receptor alpha |
| chr8_+_26240414 | 0.06 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chrY_-_2655644 | 0.06 |
ENST00000525526.2 ENST00000534739.2 ENST00000383070.1 |
SRY |
sex determining region Y |
| chr2_+_62900986 | 0.06 |
ENST00000405015.3 ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1 |
EH domain binding protein 1 |
| chr2_+_233271546 | 0.06 |
ENST00000295453.3 |
ALPPL2 |
alkaline phosphatase, placental-like 2 |
| chr2_+_220379052 | 0.05 |
ENST00000347842.3 ENST00000358078.4 |
ASIC4 |
acid-sensing (proton-gated) ion channel family member 4 |
| chr17_+_34538310 | 0.05 |
ENST00000444414.1 ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1 |
chemokine (C-C motif) ligand 4-like 1 |
| chr1_-_54872059 | 0.05 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr15_+_77287426 | 0.05 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr16_+_20817761 | 0.05 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr16_+_67360712 | 0.05 |
ENST00000569499.2 ENST00000329956.6 ENST00000561948.1 |
LRRC36 |
leucine rich repeat containing 36 |
| chr5_+_71014990 | 0.05 |
ENST00000296777.4 |
CARTPT |
CART prepropeptide |
| chr1_+_168250194 | 0.05 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr2_-_61697862 | 0.05 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr17_-_5015129 | 0.05 |
ENST00000575898.1 ENST00000416429.2 |
ZNF232 |
zinc finger protein 232 |
| chr18_+_76829385 | 0.04 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chr3_+_49726932 | 0.04 |
ENST00000327697.6 ENST00000432042.1 ENST00000454491.1 |
RNF123 |
ring finger protein 123 |
| chr16_+_1290725 | 0.04 |
ENST00000461509.2 |
TPSAB1 |
tryptase alpha/beta 1 |
| chr3_-_149093499 | 0.04 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr17_+_73089382 | 0.04 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr16_+_20817839 | 0.04 |
ENST00000348433.6 ENST00000568501.1 ENST00000566276.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr1_+_17559776 | 0.04 |
ENST00000537499.1 ENST00000413717.2 ENST00000536552.1 |
PADI1 |
peptidyl arginine deiminase, type I |
| chr22_+_46731596 | 0.04 |
ENST00000381019.3 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr3_-_130745571 | 0.04 |
ENST00000514044.1 ENST00000264992.3 |
ASTE1 |
asteroid homolog 1 (Drosophila) |
| chr12_-_54978086 | 0.04 |
ENST00000553113.1 |
PPP1R1A |
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr13_+_48877895 | 0.04 |
ENST00000267163.4 |
RB1 |
retinoblastoma 1 |
| chr11_-_82745238 | 0.03 |
ENST00000531021.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr17_-_42345487 | 0.03 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr7_+_116654935 | 0.03 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr11_+_45868957 | 0.03 |
ENST00000443527.2 |
CRY2 |
cryptochrome 2 (photolyase-like) |
| chr9_+_100174344 | 0.03 |
ENST00000422139.2 |
TDRD7 |
tudor domain containing 7 |
| chr15_-_74284613 | 0.03 |
ENST00000316911.6 ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1 |
stomatin (EPB72)-like 1 |
| chr16_+_20818020 | 0.03 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr8_+_42552533 | 0.03 |
ENST00000289957.2 |
CHRNB3 |
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr17_+_33448593 | 0.03 |
ENST00000158009.5 |
FNDC8 |
fibronectin type III domain containing 8 |
| chr19_-_4670345 | 0.02 |
ENST00000599630.1 ENST00000262947.3 |
C19orf10 |
chromosome 19 open reading frame 10 |
| chr10_-_73848764 | 0.02 |
ENST00000317376.4 ENST00000412663.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_-_155162658 | 0.02 |
ENST00000368389.2 ENST00000368396.4 ENST00000343256.5 ENST00000342482.4 ENST00000368398.3 ENST00000368390.3 ENST00000337604.5 ENST00000368392.3 ENST00000438413.1 ENST00000368393.3 ENST00000457295.2 ENST00000338684.5 ENST00000368395.1 |
MUC1 |
mucin 1, cell surface associated |
| chr3_-_133748913 | 0.02 |
ENST00000310926.4 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
| chr9_-_99540328 | 0.02 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr10_-_79398127 | 0.02 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr19_-_40791211 | 0.01 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr19_-_54850417 | 0.01 |
ENST00000291759.4 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr10_-_73848531 | 0.01 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_+_92631709 | 0.01 |
ENST00000413330.1 ENST00000277882.3 |
RPP30 |
ribonuclease P/MRP 30kDa subunit |
| chr16_+_1290694 | 0.01 |
ENST00000338844.3 |
TPSAB1 |
tryptase alpha/beta 1 |
| chr11_+_102188224 | 0.01 |
ENST00000263464.3 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr2_-_89597542 | 0.01 |
ENST00000465170.1 |
IGKV1-37 |
immunoglobulin kappa variable 1-37 (non-functional) |
| chr22_+_21369316 | 0.01 |
ENST00000413302.2 ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6 |
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr20_-_45142154 | 0.01 |
ENST00000347606.4 ENST00000457685.2 |
ZNF334 |
zinc finger protein 334 |
| chr11_-_59950519 | 0.01 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr7_+_66205712 | 0.01 |
ENST00000451741.2 ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7 RABGEF1 |
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr18_+_55018044 | 0.00 |
ENST00000324000.3 |
ST8SIA3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr20_-_3644046 | 0.00 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr22_+_42229100 | 0.00 |
ENST00000361204.4 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
| chr15_+_85427903 | 0.00 |
ENST00000286749.3 ENST00000394573.1 ENST00000537703.1 |
SLC28A1 |
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr11_-_85779971 | 0.00 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr9_+_137967268 | 0.00 |
ENST00000371799.4 ENST00000277415.11 |
OLFM1 |
olfactomedin 1 |
| chr9_+_131843377 | 0.00 |
ENST00000372546.4 ENST00000406974.3 ENST00000540102.1 |
DOLPP1 |
dolichyldiphosphatase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 1.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 1.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 1.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.3 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.3 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.5 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.8 | GO:0042118 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) endothelial cell activation(GO:0042118) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.9 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:0099541 | retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.6 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.3 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.0 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.0 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.3 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.8 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |