Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MLX_USF2_USF1_PAX2
Z-value: 0.74
Transcription factors associated with MLX_USF2_USF1_PAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
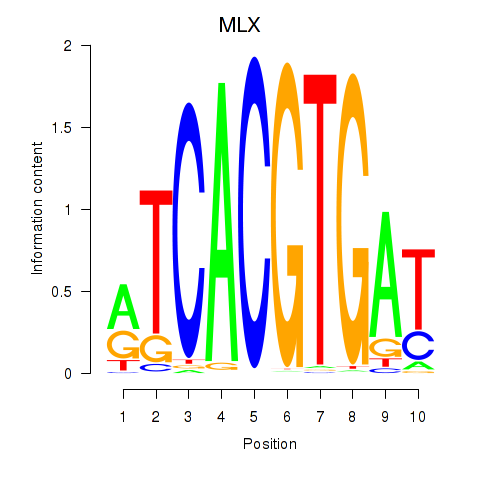
MLX
|
ENSG00000108788.7 | MLX |
|
USF2
|
ENSG00000105698.11 | USF2 |
|
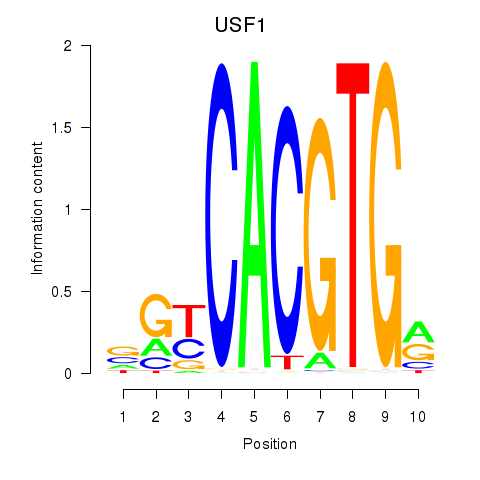
USF1
|
ENSG00000158773.10 | USF1 |
|
PAX2
|
ENSG00000075891.17 | PAX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| USF2 | hg19_v2_chr19_+_35759968_35760042 | 0.49 | 5.2e-02 | Click! |
| MLX | hg19_v2_chr17_+_40719073_40719092 | 0.44 | 9.1e-02 | Click! |
| PAX2 | hg19_v2_chr10_+_102505468_102505546 | 0.32 | 2.3e-01 | Click! |
Activity profile of MLX_USF2_USF1_PAX2 motif
Sorted Z-values of MLX_USF2_USF1_PAX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MLX_USF2_USF1_PAX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_212873267 | 1.80 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr12_+_56109926 | 1.63 |
ENST00000547076.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chrX_+_128913906 | 1.50 |
ENST00000356892.3 |
SASH3 |
SAM and SH3 domain containing 3 |
| chr12_+_56110247 | 1.49 |
ENST00000551926.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr19_+_1067271 | 1.36 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr19_+_1067144 | 1.33 |
ENST00000313093.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr1_+_183605200 | 1.30 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr19_+_1067492 | 1.30 |
ENST00000586866.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr12_+_56109810 | 1.12 |
ENST00000550412.1 ENST00000257899.2 ENST00000548925.1 ENST00000549147.1 |
RP11-644F5.10 BLOC1S1 |
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr12_-_110434096 | 1.11 |
ENST00000320063.9 ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr12_-_58146128 | 1.02 |
ENST00000551800.1 ENST00000549606.1 ENST00000257904.6 |
CDK4 |
cyclin-dependent kinase 4 |
| chr17_+_5185552 | 1.02 |
ENST00000262477.6 ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
| chr2_+_46926048 | 1.01 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr15_-_83316254 | 1.00 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr12_-_58146048 | 0.99 |
ENST00000547281.1 ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4 |
cyclin-dependent kinase 4 |
| chr7_+_116593568 | 0.94 |
ENST00000446490.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr6_-_109703600 | 0.94 |
ENST00000512821.1 |
CD164 |
CD164 molecule, sialomucin |
| chr22_-_39268308 | 0.94 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr11_+_7534999 | 0.92 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_93964158 | 0.91 |
ENST00000549206.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr6_-_33385902 | 0.90 |
ENST00000374500.5 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_111746966 | 0.90 |
ENST00000369752.5 |
DENND2D |
DENN/MADD domain containing 2D |
| chr10_-_98031310 | 0.90 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr12_+_7023491 | 0.90 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr6_-_90121938 | 0.89 |
ENST00000369415.4 |
RRAGD |
Ras-related GTP binding D |
| chr10_-_98031265 | 0.87 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr8_+_98656336 | 0.86 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr15_-_72668185 | 0.84 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr10_+_69644404 | 0.84 |
ENST00000212015.6 |
SIRT1 |
sirtuin 1 |
| chr6_-_90121789 | 0.84 |
ENST00000359203.3 |
RRAGD |
Ras-related GTP binding D |
| chr17_-_46688334 | 0.83 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr12_+_7023735 | 0.83 |
ENST00000538763.1 ENST00000544774.1 ENST00000545045.2 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr8_-_95274536 | 0.82 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr17_-_7137582 | 0.82 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr7_+_116593433 | 0.81 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr5_+_43603229 | 0.81 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr8_+_98656693 | 0.81 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr1_-_11866034 | 0.81 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr12_+_93964746 | 0.80 |
ENST00000536696.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr1_+_33116743 | 0.79 |
ENST00000414241.3 ENST00000373493.5 |
RBBP4 |
retinoblastoma binding protein 4 |
| chr19_+_50922187 | 0.79 |
ENST00000595883.1 ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr7_+_99775366 | 0.79 |
ENST00000394018.2 ENST00000416412.1 |
STAG3 |
stromal antigen 3 |
| chr18_+_9136758 | 0.79 |
ENST00000383440.2 ENST00000262126.4 ENST00000577992.1 |
ANKRD12 |
ankyrin repeat domain 12 |
| chr1_-_241520385 | 0.78 |
ENST00000366564.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr9_-_19127474 | 0.78 |
ENST00000380465.3 ENST00000380464.3 ENST00000411567.1 ENST00000276914.2 |
PLIN2 |
perilipin 2 |
| chr6_+_32811885 | 0.78 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr15_-_83316087 | 0.78 |
ENST00000568757.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr19_-_49137790 | 0.77 |
ENST00000599385.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr10_-_14050522 | 0.76 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr2_-_197036289 | 0.76 |
ENST00000263955.4 |
STK17B |
serine/threonine kinase 17b |
| chr6_+_160390102 | 0.76 |
ENST00000356956.1 |
IGF2R |
insulin-like growth factor 2 receptor |
| chrX_+_77166172 | 0.76 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_+_33116765 | 0.75 |
ENST00000544435.1 ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4 |
retinoblastoma binding protein 4 |
| chrX_-_15872914 | 0.75 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr19_+_1275917 | 0.75 |
ENST00000469144.1 |
C19orf24 |
chromosome 19 open reading frame 24 |
| chr7_+_99775520 | 0.73 |
ENST00000317296.5 ENST00000422690.1 ENST00000439782.1 |
STAG3 |
stromal antigen 3 |
| chr1_-_241520525 | 0.73 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr15_-_72668805 | 0.72 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr12_+_56110315 | 0.72 |
ENST00000548556.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr13_+_42846272 | 0.72 |
ENST00000025301.2 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
| chr5_+_126112794 | 0.72 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr16_-_89043377 | 0.72 |
ENST00000436887.2 ENST00000448839.1 ENST00000360302.2 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr16_+_2570340 | 0.72 |
ENST00000568263.1 ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2 |
amidohydrolase domain containing 2 |
| chr6_-_43337180 | 0.68 |
ENST00000318149.3 ENST00000361428.2 |
ZNF318 |
zinc finger protein 318 |
| chr14_+_75988851 | 0.67 |
ENST00000555504.1 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr18_+_32556892 | 0.67 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr6_-_33385854 | 0.67 |
ENST00000488478.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_51425902 | 0.67 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr6_-_33385870 | 0.65 |
ENST00000488034.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385823 | 0.65 |
ENST00000494751.1 ENST00000374496.3 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr6_+_13272904 | 0.65 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr6_+_87865262 | 0.65 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr2_-_158300556 | 0.64 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr22_-_39268192 | 0.64 |
ENST00000216083.6 |
CBX6 |
chromobox homolog 6 |
| chr1_-_202896310 | 0.63 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr4_-_47465666 | 0.63 |
ENST00000381571.4 |
COMMD8 |
COMM domain containing 8 |
| chr5_+_122110691 | 0.62 |
ENST00000379516.2 ENST00000505934.1 ENST00000514949.1 |
SNX2 |
sorting nexin 2 |
| chr11_+_63706444 | 0.62 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr17_-_42200996 | 0.61 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr10_+_70480963 | 0.61 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr6_-_109703663 | 0.61 |
ENST00000368961.5 |
CD164 |
CD164 molecule, sialomucin |
| chr16_-_89043605 | 0.61 |
ENST00000268679.4 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr9_-_34048873 | 0.60 |
ENST00000449054.1 ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2 |
ubiquitin associated protein 2 |
| chr19_-_48018203 | 0.60 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr3_+_127317066 | 0.59 |
ENST00000265056.7 |
MCM2 |
minichromosome maintenance complex component 2 |
| chr19_-_45681482 | 0.58 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr19_-_49137762 | 0.58 |
ENST00000593500.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr2_-_220042825 | 0.57 |
ENST00000409789.1 |
CNPPD1 |
cyclin Pas1/PHO80 domain containing 1 |
| chr3_+_113465866 | 0.57 |
ENST00000273398.3 ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr1_-_51425772 | 0.56 |
ENST00000371778.4 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr8_-_120685608 | 0.56 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr14_-_20923195 | 0.56 |
ENST00000206542.4 |
OSGEP |
O-sialoglycoprotein endopeptidase |
| chr4_-_103266355 | 0.56 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr19_-_41256207 | 0.55 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr22_+_23243156 | 0.55 |
ENST00000390323.2 |
IGLC2 |
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr20_+_44520009 | 0.55 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr17_-_35969409 | 0.54 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr2_+_143886877 | 0.54 |
ENST00000295095.6 |
ARHGAP15 |
Rho GTPase activating protein 15 |
| chr17_-_76124711 | 0.54 |
ENST00000306591.7 ENST00000590602.1 |
TMC6 |
transmembrane channel-like 6 |
| chr9_-_98279241 | 0.53 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr17_-_42200958 | 0.53 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr5_-_131132658 | 0.53 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr6_+_138188551 | 0.53 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr15_+_33022885 | 0.53 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr8_-_71581377 | 0.53 |
ENST00000276590.4 ENST00000522447.1 |
LACTB2 |
lactamase, beta 2 |
| chr2_+_216176761 | 0.52 |
ENST00000540518.1 ENST00000435675.1 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr3_+_133118839 | 0.52 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr5_+_110074685 | 0.52 |
ENST00000355943.3 ENST00000447245.2 |
SLC25A46 |
solute carrier family 25, member 46 |
| chr2_+_216176540 | 0.52 |
ENST00000236959.9 |
ATIC |
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr6_-_144329531 | 0.52 |
ENST00000429150.1 ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr2_-_175499294 | 0.52 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr6_-_33385655 | 0.51 |
ENST00000440279.3 ENST00000607266.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr1_+_40627038 | 0.51 |
ENST00000372771.4 |
RLF |
rearranged L-myc fusion |
| chr1_-_68962805 | 0.51 |
ENST00000370966.5 |
DEPDC1 |
DEP domain containing 1 |
| chr4_-_100242549 | 0.51 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr1_-_78149041 | 0.51 |
ENST00000414381.1 ENST00000370798.1 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
| chr3_-_17783990 | 0.50 |
ENST00000429383.4 ENST00000446863.1 ENST00000414349.1 ENST00000428355.1 ENST00000425944.1 ENST00000445294.1 ENST00000444471.1 ENST00000415814.2 |
TBC1D5 |
TBC1 domain family, member 5 |
| chr17_-_76124812 | 0.50 |
ENST00000592063.1 ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6 |
transmembrane channel-like 6 |
| chr10_+_52751010 | 0.50 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr5_+_43602750 | 0.50 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr16_+_81812863 | 0.50 |
ENST00000359376.3 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr17_-_7137857 | 0.49 |
ENST00000005340.5 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr1_+_145611010 | 0.49 |
ENST00000369291.5 |
RNF115 |
ring finger protein 115 |
| chr6_-_109703634 | 0.48 |
ENST00000324953.5 ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164 |
CD164 molecule, sialomucin |
| chr5_+_133706865 | 0.48 |
ENST00000265339.2 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr14_+_64970662 | 0.48 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr1_-_11865982 | 0.47 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr7_+_150065879 | 0.47 |
ENST00000397281.2 ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1 ZNF775 |
replication initiator 1 zinc finger protein 775 |
| chr12_-_109027643 | 0.47 |
ENST00000388962.3 ENST00000550948.1 |
SELPLG |
selectin P ligand |
| chr20_+_44519948 | 0.47 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chr14_-_35591433 | 0.47 |
ENST00000261475.5 ENST00000555644.1 |
PPP2R3C |
protein phosphatase 2, regulatory subunit B'', gamma |
| chr16_+_85061367 | 0.47 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr7_-_99699538 | 0.46 |
ENST00000343023.6 ENST00000303887.5 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr2_+_42396472 | 0.45 |
ENST00000318522.5 ENST00000402711.2 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr11_+_65479462 | 0.45 |
ENST00000377046.3 ENST00000352980.4 ENST00000341318.4 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr13_+_49684445 | 0.45 |
ENST00000398316.3 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr19_+_14544099 | 0.45 |
ENST00000242783.6 ENST00000586557.1 ENST00000590097.1 |
PKN1 |
protein kinase N1 |
| chr8_-_103424916 | 0.45 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr11_+_65479702 | 0.45 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr5_-_133706695 | 0.45 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr12_+_117176090 | 0.45 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr19_-_51875523 | 0.44 |
ENST00000593572.1 ENST00000595157.1 |
NKG7 |
natural killer cell group 7 sequence |
| chr10_+_112327425 | 0.44 |
ENST00000361804.4 |
SMC3 |
structural maintenance of chromosomes 3 |
| chr4_-_103266626 | 0.44 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_27961720 | 0.44 |
ENST00000545953.1 ENST00000374005.3 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr1_-_183604794 | 0.44 |
ENST00000367534.1 ENST00000359856.6 ENST00000294742.6 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr4_+_128554081 | 0.44 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr3_+_140660743 | 0.44 |
ENST00000453248.2 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr16_+_88923494 | 0.44 |
ENST00000567895.1 ENST00000301021.3 ENST00000565504.1 ENST00000567312.1 ENST00000568583.1 ENST00000561840.1 |
TRAPPC2L |
trafficking protein particle complex 2-like |
| chr21_-_46237883 | 0.44 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr11_+_95523823 | 0.44 |
ENST00000538658.1 |
CEP57 |
centrosomal protein 57kDa |
| chr3_+_112280857 | 0.43 |
ENST00000492406.1 ENST00000468642.1 |
SLC35A5 |
solute carrier family 35, member A5 |
| chr19_-_15236173 | 0.42 |
ENST00000527093.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr1_+_9599540 | 0.42 |
ENST00000302692.6 |
SLC25A33 |
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr19_-_5719860 | 0.42 |
ENST00000590729.1 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr16_+_23847339 | 0.42 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr6_-_32811771 | 0.42 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_+_127588020 | 0.42 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr1_-_31845914 | 0.42 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr12_+_117176113 | 0.42 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr14_-_64970494 | 0.42 |
ENST00000608382.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr17_-_66951474 | 0.42 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr19_-_11688500 | 0.41 |
ENST00000433365.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr8_-_103425047 | 0.41 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr11_+_75526212 | 0.41 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr6_-_151773232 | 0.41 |
ENST00000444024.1 ENST00000367303.4 |
RMND1 |
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr5_+_65440032 | 0.41 |
ENST00000334121.6 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
| chr19_-_11688447 | 0.41 |
ENST00000590420.1 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr3_-_98241358 | 0.41 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr5_-_150138551 | 0.40 |
ENST00000446090.2 ENST00000447998.2 |
DCTN4 |
dynactin 4 (p62) |
| chr19_+_56166360 | 0.40 |
ENST00000308924.4 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
| chr2_+_46926326 | 0.40 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr16_-_67517716 | 0.40 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr2_+_33701286 | 0.39 |
ENST00000403687.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_-_103424986 | 0.39 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr10_-_64576105 | 0.39 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr12_+_12870055 | 0.39 |
ENST00000228872.4 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr7_+_116593292 | 0.39 |
ENST00000393446.2 ENST00000265437.5 ENST00000393451.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr22_+_35776828 | 0.38 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr8_+_74903580 | 0.38 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr2_+_220042933 | 0.38 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chr14_-_55369525 | 0.37 |
ENST00000543643.2 ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1 |
GTP cyclohydrolase 1 |
| chr3_+_140660634 | 0.37 |
ENST00000446041.2 ENST00000507429.1 ENST00000324194.6 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr12_+_72148614 | 0.37 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr3_-_182880541 | 0.37 |
ENST00000470251.1 ENST00000265598.3 |
LAMP3 |
lysosomal-associated membrane protein 3 |
| chr12_-_123849374 | 0.37 |
ENST00000602398.1 ENST00000602750.1 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
| chr7_-_100808843 | 0.36 |
ENST00000249330.2 |
VGF |
VGF nerve growth factor inducible |
| chr21_-_46238034 | 0.36 |
ENST00000332859.6 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr2_+_74781828 | 0.36 |
ENST00000340004.6 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr4_-_76439483 | 0.36 |
ENST00000380840.2 ENST00000513257.1 ENST00000507014.1 |
RCHY1 |
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_-_198299726 | 0.36 |
ENST00000409915.4 ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1 |
splicing factor 3b, subunit 1, 155kDa |
| chr1_-_28559502 | 0.35 |
ENST00000263697.4 |
DNAJC8 |
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr1_-_111743285 | 0.35 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chr6_+_127587755 | 0.35 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr19_-_5720123 | 0.35 |
ENST00000587365.1 ENST00000585374.1 ENST00000593119.1 |
LONP1 |
lon peptidase 1, mitochondrial |
| chr8_-_102803163 | 0.35 |
ENST00000523645.1 ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD |
neurocalcin delta |
| chrX_+_131157322 | 0.35 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr9_+_6757634 | 0.34 |
ENST00000543771.1 ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C |
lysine (K)-specific demethylase 4C |
| chr14_+_75988768 | 0.34 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr19_-_51875894 | 0.34 |
ENST00000600427.1 ENST00000595217.1 ENST00000221978.5 |
NKG7 |
natural killer cell group 7 sequence |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 1.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.3 | 1.0 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 1.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 1.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 0.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.8 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 1.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.9 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.0 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.3 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 2.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.2 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0035242 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 2.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 2.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 2.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 1.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 5.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 1.6 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 0.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 0.6 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 1.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.2 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.3 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 1.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 1.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 3.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.4 | 1.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 1.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.4 | 2.0 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.4 | 1.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.3 | 1.0 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.3 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.3 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.7 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 0.6 | GO:1904029 | regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.3 | 0.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 0.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.3 | 1.3 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.3 | 0.8 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 0.2 | GO:0045346 | MHC class II biosynthetic process(GO:0045342) regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 1.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.6 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.2 | 1.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.2 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 0.5 | GO:0002316 | follicular B cell differentiation(GO:0002316) activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 0.6 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.2 | 0.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 1.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:0021569 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 1.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.5 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.5 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 1.0 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.3 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.3 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 0.6 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 2.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.4 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.2 | GO:0072171 | branching involved in ureteric bud morphogenesis(GO:0001658) ureteric bud morphogenesis(GO:0060675) mesonephric tubule morphogenesis(GO:0072171) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.2 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.4 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.9 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.5 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.6 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.4 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.0 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.5 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0070486 | leukocyte aggregation(GO:0070486) |
| 0.0 | 0.1 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 2.0 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.7 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.4 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.6 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:1900125 | regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.4 | GO:0033008 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0046098 | hypoxanthine salvage(GO:0043103) guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:1900040 | regulation of interleukin-2 secretion(GO:1900040) positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:2000104 | negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 1.3 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.0 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0045162 | skeletal muscle hypertrophy(GO:0014734) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.7 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.2 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.1 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.0 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:1902739 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.0 | GO:0035822 | gene conversion(GO:0035822) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0046066 | dGDP metabolic process(GO:0046066) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.1 | GO:0060748 | paracrine signaling(GO:0038001) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 1.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.0 | GO:0007619 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.6 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.0 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.1 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.6 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |