Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
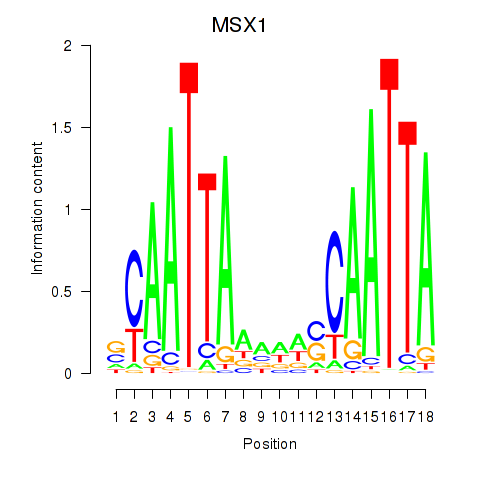
Results for MSX1
Z-value: 0.99
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | MSX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg19_v2_chr4_+_4861385_4861398 | -0.29 | 2.7e-01 | Click! |
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_38172863 | 8.49 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr1_+_158985457 | 3.57 |
ENST00000567661.1 ENST00000474473.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr14_+_55595762 | 3.37 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr8_+_30244580 | 2.90 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr14_+_55595960 | 2.73 |
ENST00000554715.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr3_-_123339418 | 2.58 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr3_-_123339343 | 2.52 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr3_-_123512688 | 2.30 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr18_+_61637159 | 1.61 |
ENST00000397985.2 ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr21_+_30502806 | 1.54 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr12_-_52911718 | 1.51 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr9_+_79074068 | 1.41 |
ENST00000444201.2 ENST00000376730.4 |
GCNT1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr2_-_31361543 | 1.36 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr5_+_82767487 | 1.35 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_+_82767284 | 1.09 |
ENST00000265077.3 |
VCAN |
versican |
| chr10_+_13141585 | 1.05 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr9_+_115913222 | 1.05 |
ENST00000259392.3 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
| chr5_+_82767583 | 0.99 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr11_+_12766583 | 0.99 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr10_+_13142225 | 0.98 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr5_+_140762268 | 0.94 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr10_+_17270214 | 0.90 |
ENST00000544301.1 |
VIM |
vimentin |
| chr16_-_66584059 | 0.89 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr12_+_56324756 | 0.82 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr6_-_109777128 | 0.73 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr4_-_57547454 | 0.73 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr1_-_149908217 | 0.66 |
ENST00000369140.3 |
MTMR11 |
myotubularin related protein 11 |
| chr11_+_12132117 | 0.65 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr1_+_84630645 | 0.62 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_224370873 | 0.61 |
ENST00000323699.4 ENST00000391877.3 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
| chr10_-_101380121 | 0.59 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr7_-_32338917 | 0.51 |
ENST00000396193.1 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr1_-_35450897 | 0.48 |
ENST00000373337.3 |
ZMYM6NB |
ZMYM6 neighbor |
| chr22_+_51176624 | 0.46 |
ENST00000216139.5 ENST00000529621.1 |
ACR |
acrosin |
| chr14_-_64194745 | 0.46 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr5_+_67586465 | 0.45 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr14_+_22964877 | 0.44 |
ENST00000390494.1 |
TRAJ43 |
T cell receptor alpha joining 43 |
| chr4_+_169013666 | 0.41 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr16_-_66583701 | 0.38 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chrX_+_1710484 | 0.37 |
ENST00000313871.3 ENST00000381261.3 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
| chr6_-_32557610 | 0.37 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr5_+_150404904 | 0.36 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr5_+_175490540 | 0.35 |
ENST00000515817.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr12_+_54422142 | 0.34 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr12_+_56324933 | 0.34 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr3_+_93781728 | 0.32 |
ENST00000314622.4 |
NSUN3 |
NOP2/Sun domain family, member 3 |
| chr1_+_210501589 | 0.30 |
ENST00000413764.2 ENST00000541565.1 |
HHAT |
hedgehog acyltransferase |
| chr10_+_135204338 | 0.30 |
ENST00000468317.2 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
| chr11_-_85376121 | 0.30 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chr22_-_32766972 | 0.28 |
ENST00000382084.4 ENST00000382086.2 |
RFPL3S |
RFPL3 antisense |
| chr6_-_32784687 | 0.28 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr5_-_177207634 | 0.26 |
ENST00000513554.1 ENST00000440605.3 |
FAM153A |
family with sequence similarity 153, member A |
| chr2_-_136288113 | 0.26 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr20_+_34129770 | 0.25 |
ENST00000348547.2 ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3 |
ERGIC and golgi 3 |
| chr6_+_6588316 | 0.25 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr6_-_32095968 | 0.24 |
ENST00000375203.3 ENST00000375201.4 |
ATF6B |
activating transcription factor 6 beta |
| chr7_-_16840820 | 0.23 |
ENST00000450569.1 |
AGR2 |
anterior gradient 2 |
| chr18_+_29598335 | 0.23 |
ENST00000217740.3 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
| chr8_+_97597148 | 0.23 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chrX_+_73164149 | 0.22 |
ENST00000602938.1 ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX |
JPX transcript, XIST activator (non-protein coding) |
| chr5_-_137475071 | 0.21 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chr6_-_52705641 | 0.21 |
ENST00000370989.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr3_+_108308513 | 0.19 |
ENST00000361582.3 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr8_-_99057742 | 0.19 |
ENST00000521291.1 ENST00000396070.2 ENST00000523172.1 ENST00000287038.3 |
RPL30 |
ribosomal protein L30 |
| chr15_-_77712477 | 0.19 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr15_-_42500351 | 0.19 |
ENST00000348544.4 ENST00000318006.5 |
VPS39 |
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr13_+_50570019 | 0.18 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr15_+_79166065 | 0.18 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr1_+_160160283 | 0.17 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_+_140220769 | 0.16 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chrX_+_11311533 | 0.16 |
ENST00000380714.3 ENST00000380712.3 ENST00000348912.4 |
AMELX |
amelogenin, X-linked |
| chr4_-_120243545 | 0.15 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr14_+_22293618 | 0.15 |
ENST00000390432.2 |
TRAV10 |
T cell receptor alpha variable 10 |
| chr1_+_160160346 | 0.13 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr13_+_49551020 | 0.12 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr22_-_32767017 | 0.12 |
ENST00000400234.1 |
RFPL3S |
RFPL3 antisense |
| chr8_+_24241789 | 0.12 |
ENST00000256412.4 ENST00000538205.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr1_-_167487758 | 0.11 |
ENST00000362089.5 |
CD247 |
CD247 molecule |
| chr8_-_17555164 | 0.10 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr11_-_128894053 | 0.10 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr6_+_29141311 | 0.09 |
ENST00000377167.2 |
OR2J2 |
olfactory receptor, family 2, subfamily J, member 2 |
| chr19_-_53758094 | 0.09 |
ENST00000601828.1 ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677 |
zinc finger protein 677 |
| chr11_-_66964638 | 0.09 |
ENST00000444002.2 |
AP001885.1 |
AP001885.1 |
| chr8_-_121824374 | 0.09 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr21_+_43619796 | 0.08 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr1_+_158815588 | 0.06 |
ENST00000438394.1 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr7_+_2636522 | 0.06 |
ENST00000423196.1 |
IQCE |
IQ motif containing E |
| chr1_-_167487808 | 0.06 |
ENST00000392122.3 |
CD247 |
CD247 molecule |
| chr19_-_20150014 | 0.05 |
ENST00000358523.5 ENST00000397162.1 ENST00000601100.1 |
ZNF682 |
zinc finger protein 682 |
| chr10_+_135050908 | 0.04 |
ENST00000325980.9 |
VENTX |
VENT homeobox |
| chr19_+_55043977 | 0.04 |
ENST00000335056.3 |
KIR3DX1 |
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr2_-_219031709 | 0.04 |
ENST00000295683.2 |
CXCR1 |
chemokine (C-X-C motif) receptor 1 |
| chr1_+_202091980 | 0.04 |
ENST00000367282.5 |
GPR37L1 |
G protein-coupled receptor 37 like 1 |
| chr17_-_37934466 | 0.03 |
ENST00000583368.1 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
| chr3_-_190040223 | 0.03 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr8_+_24241969 | 0.02 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr2_-_158345462 | 0.02 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr11_+_45825616 | 0.02 |
ENST00000442528.2 ENST00000456334.1 ENST00000526817.1 |
SLC35C1 |
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr14_-_75389975 | 0.01 |
ENST00000555647.1 ENST00000557413.1 |
RPS6KL1 |
ribosomal protein S6 kinase-like 1 |
| chr6_-_84419101 | 0.00 |
ENST00000520302.1 ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91 |
synaptosomal-associated protein, 91kDa |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 7.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 7.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 8.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 3.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 2.0 | 6.1 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.9 | 7.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 1.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 1.3 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 2.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 3.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 2.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 1.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 1.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.6 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 3.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 1.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:1990440 | ATF6-mediated unfolded protein response(GO:0036500) positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.8 | 6.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.5 | 1.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.4 | 1.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.4 | 2.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.5 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.1 | 0.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 2.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 1.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 8.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 3.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 6.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 8.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |