Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MSX2
Z-value: 0.43
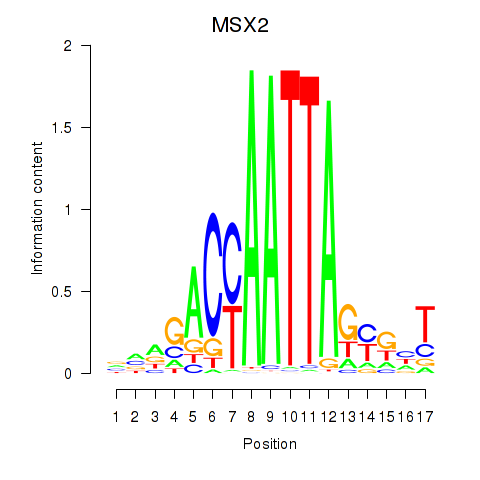
Transcription factors associated with MSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX2
|
ENSG00000120149.7 | MSX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX2 | hg19_v2_chr5_+_174151536_174151610 | 0.59 | 1.5e-02 | Click! |
Activity profile of MSX2 motif
Sorted Z-values of MSX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_151965048 | 1.78 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr1_-_68698222 | 1.12 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr6_-_167040731 | 1.02 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr1_-_94050668 | 0.89 |
ENST00000539242.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr3_-_151034734 | 0.78 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr1_-_68698197 | 0.66 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr12_-_10022735 | 0.60 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chrX_-_48931648 | 0.48 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr12_+_9102632 | 0.47 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr3_-_112127981 | 0.47 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr5_+_95997918 | 0.47 |
ENST00000395812.2 ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST |
calpastatin |
| chr7_+_134331550 | 0.45 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr6_-_111927449 | 0.40 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr5_+_66254698 | 0.38 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_95998070 | 0.37 |
ENST00000421689.2 ENST00000510756.1 ENST00000512620.1 |
CAST |
calpastatin |
| chr5_+_140797296 | 0.35 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr10_-_99447024 | 0.35 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr15_+_41245160 | 0.35 |
ENST00000444189.2 ENST00000446533.3 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr7_-_2883928 | 0.33 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chr6_-_111927062 | 0.33 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr7_+_100210133 | 0.31 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr7_+_100209725 | 0.29 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr7_+_100209979 | 0.29 |
ENST00000493970.1 ENST00000379527.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr1_+_44115814 | 0.21 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr12_+_54718904 | 0.21 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chr12_-_9102549 | 0.20 |
ENST00000000412.3 |
M6PR |
mannose-6-phosphate receptor (cation dependent) |
| chr1_-_21377383 | 0.19 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr4_-_69536346 | 0.19 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_-_151762943 | 0.18 |
ENST00000368825.3 ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH |
tudor and KH domain containing |
| chr9_+_34646651 | 0.18 |
ENST00000378842.3 |
GALT |
galactose-1-phosphate uridylyltransferase |
| chr1_+_151735431 | 0.17 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr9_+_34646624 | 0.17 |
ENST00000450095.2 ENST00000556278.1 |
GALT GALT |
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr4_+_71587669 | 0.16 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr7_-_100183742 | 0.16 |
ENST00000310300.6 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr10_-_28571015 | 0.14 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_+_9413410 | 0.12 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr20_+_54967409 | 0.12 |
ENST00000415828.1 ENST00000217109.4 ENST00000428552.1 |
CSTF1 |
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chrX_-_139587225 | 0.10 |
ENST00000370536.2 |
SOX3 |
SRY (sex determining region Y)-box 3 |
| chr7_+_100183927 | 0.10 |
ENST00000241071.6 ENST00000360609.2 |
FBXO24 |
F-box protein 24 |
| chr11_-_71823266 | 0.10 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr4_-_69434245 | 0.09 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr20_+_56964169 | 0.09 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr19_+_17378278 | 0.09 |
ENST00000596335.1 ENST00000601436.1 ENST00000595632.1 |
BABAM1 |
BRISC and BRCA1 A complex member 1 |
| chr5_-_132200477 | 0.09 |
ENST00000296875.2 |
GDF9 |
growth differentiation factor 9 |
| chr7_-_99716952 | 0.09 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_202317815 | 0.09 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr9_-_95056010 | 0.08 |
ENST00000443024.2 |
IARS |
isoleucyl-tRNA synthetase |
| chr14_-_23504337 | 0.08 |
ENST00000361611.6 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr17_-_43045439 | 0.08 |
ENST00000253407.3 |
C1QL1 |
complement component 1, q subcomponent-like 1 |
| chr5_+_150639360 | 0.08 |
ENST00000523004.1 |
GM2A |
GM2 ganglioside activator |
| chr3_-_114866084 | 0.07 |
ENST00000357258.3 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_+_17298297 | 0.07 |
ENST00000529010.1 |
NUCB2 |
nucleobindin 2 |
| chr3_-_11685345 | 0.07 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr19_+_17378159 | 0.06 |
ENST00000598188.1 ENST00000359435.4 ENST00000599474.1 ENST00000599057.1 ENST00000601043.1 ENST00000447614.2 |
BABAM1 |
BRISC and BRCA1 A complex member 1 |
| chr11_-_71823796 | 0.06 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr11_-_71823715 | 0.06 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr14_-_23504432 | 0.05 |
ENST00000425762.2 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr11_-_13517565 | 0.05 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr12_-_10955226 | 0.05 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr12_-_57039739 | 0.05 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr2_-_98280383 | 0.05 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr11_+_17298255 | 0.03 |
ENST00000531172.1 ENST00000533738.2 ENST00000323688.6 |
NUCB2 |
nucleobindin 2 |
| chrX_+_113818545 | 0.03 |
ENST00000371951.1 ENST00000276198.1 ENST00000371950.3 |
HTR2C |
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| chr19_-_10121144 | 0.03 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr4_-_46996424 | 0.03 |
ENST00000264318.3 |
GABRA4 |
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr8_+_1993173 | 0.02 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr9_-_5339873 | 0.02 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr1_-_228613026 | 0.01 |
ENST00000366696.1 |
HIST3H3 |
histone cluster 3, H3 |
| chr8_+_1993152 | 0.01 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr16_+_69345243 | 0.01 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr3_+_129247479 | 0.01 |
ENST00000296271.3 |
RHO |
rhodopsin |
| chr18_+_39535152 | 0.00 |
ENST00000262039.4 ENST00000398870.3 ENST00000586545.1 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr19_+_42817527 | 0.00 |
ENST00000598766.1 |
TMEM145 |
transmembrane protein 145 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.8 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.1 | 0.8 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |