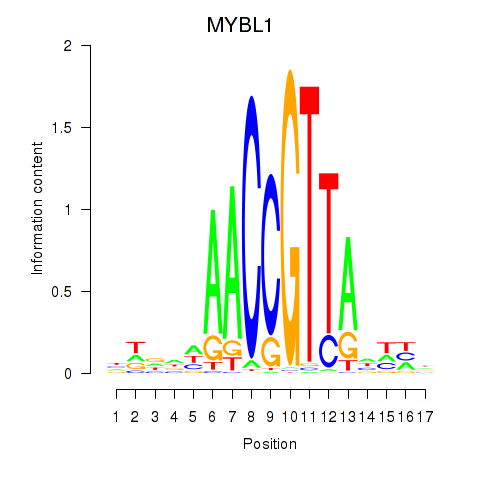
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for MYBL1
Z-value: 1.00
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYBL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525473_67525518 | 0.23 | 3.9e-01 | Click! |
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_157154578 | 4.50 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr19_-_36643329 | 2.20 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr10_+_31608054 | 1.82 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr12_-_91539918 | 1.64 |
ENST00000548218.1 |
DCN |
decorin |
| chr5_-_88179302 | 1.43 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr1_+_155829286 | 1.29 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr1_+_211499957 | 1.22 |
ENST00000336184.2 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr12_-_25055177 | 1.21 |
ENST00000538118.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr1_+_211500129 | 1.21 |
ENST00000427925.2 ENST00000261464.5 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr12_-_91576561 | 1.17 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr5_-_88179017 | 1.15 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_-_190044480 | 1.06 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_-_153517473 | 1.01 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr11_-_27723158 | 0.96 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr6_+_33043703 | 0.90 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr12_+_10365404 | 0.88 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr21_-_40033618 | 0.78 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr14_+_61654271 | 0.78 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr4_+_90800656 | 0.76 |
ENST00000394980.1 |
MMRN1 |
multimerin 1 |
| chr5_-_146833485 | 0.75 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr19_-_43708378 | 0.73 |
ENST00000599746.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr3_+_138067521 | 0.72 |
ENST00000494949.1 |
MRAS |
muscle RAS oncogene homolog |
| chr4_+_123747834 | 0.68 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr11_+_2407287 | 0.65 |
ENST00000381036.3 ENST00000492252.1 |
CD81 |
CD81 molecule |
| chr6_-_56707943 | 0.59 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr14_+_65007177 | 0.56 |
ENST00000247207.6 |
HSPA2 |
heat shock 70kDa protein 2 |
| chr1_+_97187318 | 0.56 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr15_-_63449663 | 0.56 |
ENST00000439025.1 |
RPS27L |
ribosomal protein S27-like |
| chr11_-_6677018 | 0.56 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr6_+_42847348 | 0.54 |
ENST00000493763.1 ENST00000304734.5 |
RPL7L1 |
ribosomal protein L7-like 1 |
| chr4_+_123747979 | 0.52 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr2_+_33661382 | 0.51 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr12_-_10251603 | 0.50 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr13_-_33760216 | 0.50 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr2_-_133427767 | 0.49 |
ENST00000397463.2 |
LYPD1 |
LY6/PLAUR domain containing 1 |
| chr12_-_76478446 | 0.49 |
ENST00000393263.3 ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chrX_-_104465358 | 0.48 |
ENST00000372578.3 ENST00000372575.1 ENST00000413579.1 |
TEX13A |
testis expressed 13A |
| chr18_-_71959159 | 0.46 |
ENST00000494131.2 ENST00000397914.4 ENST00000340533.4 |
CYB5A |
cytochrome b5 type A (microsomal) |
| chr3_+_141144954 | 0.46 |
ENST00000441582.2 ENST00000321464.5 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr3_+_173116225 | 0.46 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr12_-_76478686 | 0.45 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr3_-_148804275 | 0.45 |
ENST00000392912.2 ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF |
helicase-like transcription factor |
| chr11_-_47600549 | 0.45 |
ENST00000430070.2 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr11_-_47600320 | 0.44 |
ENST00000525720.1 ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr6_+_13272904 | 0.44 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr11_-_104840093 | 0.43 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr12_-_14996355 | 0.41 |
ENST00000228936.4 |
ART4 |
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr2_+_85766280 | 0.40 |
ENST00000306434.3 |
MAT2A |
methionine adenosyltransferase II, alpha |
| chr4_+_24797085 | 0.38 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr8_-_145652336 | 0.37 |
ENST00000529182.1 ENST00000526054.1 |
VPS28 |
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr21_-_43816052 | 0.37 |
ENST00000398405.1 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr12_+_56114189 | 0.37 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_-_86564776 | 0.36 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr6_-_32557610 | 0.36 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr14_-_25479811 | 0.36 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr12_+_56114151 | 0.36 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_+_65339820 | 0.35 |
ENST00000316409.2 ENST00000449319.2 ENST00000530349.1 |
FAM89B |
family with sequence similarity 89, member B |
| chrY_+_15815447 | 0.34 |
ENST00000284856.3 |
TMSB4Y |
thymosin beta 4, Y-linked |
| chr7_-_38948774 | 0.34 |
ENST00000395969.2 ENST00000414632.1 ENST00000310301.4 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr16_-_425205 | 0.34 |
ENST00000448854.1 |
TMEM8A |
transmembrane protein 8A |
| chr1_-_159046617 | 0.34 |
ENST00000368130.4 |
AIM2 |
absent in melanoma 2 |
| chr17_+_48243352 | 0.33 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr4_+_76649797 | 0.32 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr10_-_23003460 | 0.31 |
ENST00000376573.4 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr6_+_71377473 | 0.31 |
ENST00000370452.3 ENST00000316999.5 ENST00000370455.3 |
SMAP1 |
small ArfGAP 1 |
| chr22_-_36220420 | 0.30 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_153637612 | 0.30 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr14_-_106725723 | 0.29 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr6_+_74104471 | 0.29 |
ENST00000370336.4 |
DDX43 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr19_-_55919087 | 0.28 |
ENST00000587845.1 ENST00000589978.1 ENST00000264552.9 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
| chr16_-_30569584 | 0.28 |
ENST00000252797.2 ENST00000568114.1 |
ZNF764 AC002310.13 |
zinc finger protein 764 Uncharacterized protein |
| chr11_-_62323702 | 0.28 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr18_+_3247779 | 0.27 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_145076186 | 0.27 |
ENST00000369348.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr19_-_12780211 | 0.27 |
ENST00000597961.1 ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24 WDR83OS |
Uncharacterized protein WD repeat domain 83 opposite strand |
| chr9_-_116163400 | 0.27 |
ENST00000277315.5 ENST00000448137.1 ENST00000409155.3 |
ALAD |
aminolevulinate dehydratase |
| chr2_-_87248975 | 0.26 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr11_+_43380459 | 0.26 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr12_-_66563728 | 0.25 |
ENST00000286424.7 ENST00000556010.1 ENST00000398033.4 |
TMBIM4 |
transmembrane BAX inhibitor motif containing 4 |
| chr12_-_120765565 | 0.25 |
ENST00000423423.3 ENST00000308366.4 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr7_+_80275621 | 0.25 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr3_-_3221358 | 0.24 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr17_+_16284104 | 0.24 |
ENST00000577958.1 ENST00000302182.3 ENST00000577640.1 |
UBB |
ubiquitin B |
| chr19_-_8408139 | 0.24 |
ENST00000330915.3 ENST00000593649.1 ENST00000595639.1 |
KANK3 |
KN motif and ankyrin repeat domains 3 |
| chr17_-_1420182 | 0.24 |
ENST00000421807.2 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr16_+_89753070 | 0.24 |
ENST00000353379.7 ENST00000505473.1 ENST00000564192.1 |
CDK10 |
cyclin-dependent kinase 10 |
| chr21_-_15755446 | 0.23 |
ENST00000544452.1 ENST00000285667.3 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
| chr22_+_22712087 | 0.23 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chr3_+_160117418 | 0.23 |
ENST00000465903.1 ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4 |
structural maintenance of chromosomes 4 |
| chr22_-_39268308 | 0.23 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr16_-_29934558 | 0.23 |
ENST00000568995.1 ENST00000566413.1 |
KCTD13 |
potassium channel tetramerization domain containing 13 |
| chr11_+_64323098 | 0.22 |
ENST00000301891.4 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr7_+_80275752 | 0.22 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr1_-_43855444 | 0.22 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr17_-_9479128 | 0.22 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr16_-_30569801 | 0.21 |
ENST00000395091.2 |
ZNF764 |
zinc finger protein 764 |
| chr12_+_53835508 | 0.21 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr1_-_43855479 | 0.21 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr12_+_133758115 | 0.21 |
ENST00000541009.2 ENST00000592241.1 |
ZNF268 |
zinc finger protein 268 |
| chr17_+_7487146 | 0.20 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr13_-_114567034 | 0.20 |
ENST00000327773.6 ENST00000357389.3 |
GAS6 |
growth arrest-specific 6 |
| chr17_+_16284604 | 0.20 |
ENST00000395839.1 ENST00000395837.1 |
UBB |
ubiquitin B |
| chr21_-_43816152 | 0.19 |
ENST00000433957.2 ENST00000398397.3 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr10_+_71561630 | 0.19 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr1_-_84464780 | 0.19 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr15_+_81589254 | 0.18 |
ENST00000394652.2 |
IL16 |
interleukin 16 |
| chr6_+_88182643 | 0.18 |
ENST00000369556.3 ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1 |
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr13_-_37633567 | 0.18 |
ENST00000464744.1 |
SUPT20H |
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr10_+_91174314 | 0.18 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chr2_+_204193101 | 0.17 |
ENST00000430418.1 ENST00000424558.1 ENST00000261016.6 |
ABI2 |
abl-interactor 2 |
| chr13_+_32838801 | 0.17 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr2_+_88047606 | 0.17 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr2_-_190627481 | 0.17 |
ENST00000264151.5 ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1 |
O-sialoglycoprotein endopeptidase-like 1 |
| chr17_+_40172069 | 0.17 |
ENST00000393885.4 ENST00000393884.2 ENST00000587337.1 ENST00000479407.1 |
NKIRAS2 |
NFKB inhibitor interacting Ras-like 2 |
| chr1_-_19615744 | 0.17 |
ENST00000361640.4 |
AKR7A3 |
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr19_-_52551814 | 0.16 |
ENST00000594154.1 ENST00000598745.1 ENST00000597273.1 |
ZNF432 |
zinc finger protein 432 |
| chr11_+_64323156 | 0.16 |
ENST00000377585.3 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr21_-_46221684 | 0.16 |
ENST00000330942.5 |
UBE2G2 |
ubiquitin-conjugating enzyme E2G 2 |
| chr19_-_1605424 | 0.15 |
ENST00000589880.1 ENST00000585671.1 ENST00000591899.3 |
UQCR11 |
ubiquinol-cytochrome c reductase, complex III subunit XI |
| chr1_+_44115814 | 0.15 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr8_-_23315190 | 0.15 |
ENST00000356206.6 ENST00000358689.4 ENST00000417069.2 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr14_-_75518129 | 0.15 |
ENST00000556257.1 ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3 |
mutL homolog 3 |
| chr4_+_156824840 | 0.15 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr16_-_22385901 | 0.15 |
ENST00000268383.2 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
| chrX_+_148622513 | 0.15 |
ENST00000393985.3 ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr17_+_16284399 | 0.15 |
ENST00000535788.1 |
UBB |
ubiquitin B |
| chr10_-_35379524 | 0.14 |
ENST00000374751.3 ENST00000374742.1 ENST00000602371.1 |
CUL2 |
cullin 2 |
| chr11_+_93479588 | 0.14 |
ENST00000526335.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
| chr9_+_100263912 | 0.14 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chrX_+_114795489 | 0.14 |
ENST00000355899.3 ENST00000537301.1 ENST00000289290.3 |
PLS3 |
plastin 3 |
| chr18_+_3247413 | 0.14 |
ENST00000579226.1 ENST00000217652.3 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr16_+_14165160 | 0.13 |
ENST00000574998.1 ENST00000571589.1 ENST00000574045.1 |
MKL2 |
MKL/myocardin-like 2 |
| chr13_-_37633743 | 0.13 |
ENST00000497318.1 ENST00000475892.1 ENST00000356185.3 ENST00000350612.6 ENST00000542180.1 ENST00000360252.4 |
SUPT20H |
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr11_-_59950519 | 0.13 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr14_+_60558627 | 0.13 |
ENST00000317623.4 ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4 |
pecanex-like 4 (Drosophila) |
| chr6_-_39902185 | 0.13 |
ENST00000373195.3 ENST00000308559.7 ENST00000373188.2 |
MOCS1 |
molybdenum cofactor synthesis 1 |
| chr7_-_148581360 | 0.13 |
ENST00000320356.2 ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr4_+_175839551 | 0.12 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr13_+_21714653 | 0.12 |
ENST00000382533.4 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr8_-_117768023 | 0.12 |
ENST00000518949.1 ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
| chr1_-_68915610 | 0.12 |
ENST00000262340.5 |
RPE65 |
retinal pigment epithelium-specific protein 65kDa |
| chr11_-_59950622 | 0.12 |
ENST00000323961.3 ENST00000412309.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr1_+_145883868 | 0.12 |
ENST00000447947.2 |
GPR89C |
G protein-coupled receptor 89C |
| chrX_+_153627231 | 0.12 |
ENST00000406022.2 |
RPL10 |
ribosomal protein L10 |
| chr6_+_35227449 | 0.12 |
ENST00000373953.3 ENST00000440666.2 ENST00000339411.5 |
ZNF76 |
zinc finger protein 76 |
| chr17_-_1303462 | 0.11 |
ENST00000573026.1 ENST00000575977.1 ENST00000571732.1 ENST00000264335.8 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr17_+_40172097 | 0.11 |
ENST00000393880.1 ENST00000393881.3 ENST00000462043.2 ENST00000449471.4 ENST00000316082.4 |
NKIRAS2 |
NFKB inhibitor interacting Ras-like 2 |
| chr22_-_42343117 | 0.11 |
ENST00000407253.3 ENST00000215980.5 |
CENPM |
centromere protein M |
| chr7_-_16685422 | 0.11 |
ENST00000306999.2 |
ANKMY2 |
ankyrin repeat and MYND domain containing 2 |
| chr20_-_52687059 | 0.10 |
ENST00000371435.2 ENST00000395961.3 |
BCAS1 |
breast carcinoma amplified sequence 1 |
| chr4_-_122791583 | 0.10 |
ENST00000506636.1 ENST00000264499.4 |
BBS7 |
Bardet-Biedl syndrome 7 |
| chr2_+_204103663 | 0.10 |
ENST00000356079.4 ENST00000429815.2 |
CYP20A1 |
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr17_-_30668887 | 0.10 |
ENST00000581747.1 ENST00000583334.1 ENST00000580558.1 |
C17orf75 |
chromosome 17 open reading frame 75 |
| chr3_+_134204551 | 0.10 |
ENST00000332047.5 ENST00000354446.3 |
CEP63 |
centrosomal protein 63kDa |
| chr3_-_134204815 | 0.10 |
ENST00000514612.1 ENST00000510994.1 ENST00000354910.5 |
ANAPC13 |
anaphase promoting complex subunit 13 |
| chr5_-_176778803 | 0.10 |
ENST00000303127.7 |
LMAN2 |
lectin, mannose-binding 2 |
| chr8_-_133687813 | 0.10 |
ENST00000250173.1 ENST00000519595.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr11_-_59950486 | 0.09 |
ENST00000426738.2 ENST00000533023.1 ENST00000420732.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr6_-_27782548 | 0.09 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr22_-_50708781 | 0.09 |
ENST00000449719.2 ENST00000330651.6 |
MAPK11 |
mitogen-activated protein kinase 11 |
| chr14_-_78174344 | 0.09 |
ENST00000216489.3 |
ALKBH1 |
alkB, alkylation repair homolog 1 (E. coli) |
| chr1_+_172502336 | 0.09 |
ENST00000263688.3 |
SUCO |
SUN domain containing ossification factor |
| chr2_-_55646957 | 0.09 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr6_-_29343068 | 0.09 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr6_+_28193037 | 0.09 |
ENST00000531981.1 ENST00000425468.2 ENST00000252207.5 ENST00000531979.1 ENST00000527436.1 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
| chr17_+_685513 | 0.08 |
ENST00000304478.4 |
RNMTL1 |
RNA methyltransferase like 1 |
| chr3_+_134205000 | 0.08 |
ENST00000512894.1 ENST00000513612.2 ENST00000606977.1 |
CEP63 |
centrosomal protein 63kDa |
| chr1_-_160232312 | 0.08 |
ENST00000440682.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr12_+_53835383 | 0.08 |
ENST00000429243.2 |
PRR13 |
proline rich 13 |
| chr3_+_49977440 | 0.08 |
ENST00000442092.1 ENST00000266022.4 ENST00000443081.1 |
RBM6 |
RNA binding motif protein 6 |
| chr2_-_191115229 | 0.08 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chr1_+_210001309 | 0.08 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr12_+_53835539 | 0.08 |
ENST00000547368.1 ENST00000379786.4 ENST00000551945.1 |
PRR13 |
proline rich 13 |
| chr7_-_124569991 | 0.08 |
ENST00000446993.1 ENST00000357628.3 ENST00000393329.1 |
POT1 |
protection of telomeres 1 |
| chr7_-_1980128 | 0.08 |
ENST00000437877.1 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr3_+_134204881 | 0.08 |
ENST00000511574.1 ENST00000337090.3 ENST00000383229.3 |
CEP63 |
centrosomal protein 63kDa |
| chr14_+_51706886 | 0.07 |
ENST00000457354.2 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
| chr3_+_50712672 | 0.07 |
ENST00000266037.9 |
DOCK3 |
dedicator of cytokinesis 3 |
| chr6_+_36562132 | 0.07 |
ENST00000373715.6 ENST00000339436.7 |
SRSF3 |
serine/arginine-rich splicing factor 3 |
| chr7_-_123198284 | 0.07 |
ENST00000355749.2 |
NDUFA5 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr18_-_44497308 | 0.07 |
ENST00000585916.1 ENST00000324794.7 ENST00000545673.1 |
PIAS2 |
protein inhibitor of activated STAT, 2 |
| chr15_+_78730531 | 0.07 |
ENST00000258886.8 |
IREB2 |
iron-responsive element binding protein 2 |
| chr11_+_47600562 | 0.07 |
ENST00000263774.4 ENST00000529276.1 ENST00000528192.1 ENST00000530295.1 ENST00000534208.1 ENST00000534716.2 |
NDUFS3 |
NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| chr1_-_193074504 | 0.07 |
ENST00000367439.3 |
GLRX2 |
glutaredoxin 2 |
| chr2_+_201936458 | 0.07 |
ENST00000237889.4 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr1_+_43855560 | 0.07 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr11_-_6495101 | 0.07 |
ENST00000528227.1 ENST00000359518.3 ENST00000345851.3 ENST00000537602.1 |
TRIM3 |
tripartite motif containing 3 |
| chr19_+_24269981 | 0.07 |
ENST00000339642.6 ENST00000357002.4 |
ZNF254 |
zinc finger protein 254 |
| chr12_-_47219733 | 0.07 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr2_-_55647057 | 0.06 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr17_-_30669138 | 0.06 |
ENST00000225805.4 ENST00000577809.1 |
C17orf75 |
chromosome 17 open reading frame 75 |
| chr6_+_26240561 | 0.06 |
ENST00000377745.2 |
HIST1H4F |
histone cluster 1, H4f |
| chr19_-_47734448 | 0.06 |
ENST00000439096.2 |
BBC3 |
BCL2 binding component 3 |
| chr19_+_10381769 | 0.06 |
ENST00000423829.2 ENST00000588645.1 |
ICAM1 |
intercellular adhesion molecule 1 |
| chr7_-_151217166 | 0.06 |
ENST00000496004.1 |
RHEB |
Ras homolog enriched in brain |
| chr11_+_118938485 | 0.05 |
ENST00000300793.6 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr14_+_22739823 | 0.05 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr8_-_100905850 | 0.05 |
ENST00000520271.1 ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr4_-_140098339 | 0.05 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr7_-_80551671 | 0.05 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 2.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 2.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 2.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.0 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.6 | 2.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 1.3 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.3 | 1.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 1.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.2 | 2.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.8 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 0.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 1.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 0.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.4 | GO:0070086 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 1.0 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.3 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.5 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 2.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:1900113 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.4 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.4 | GO:0071450 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.9 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 3.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |