Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
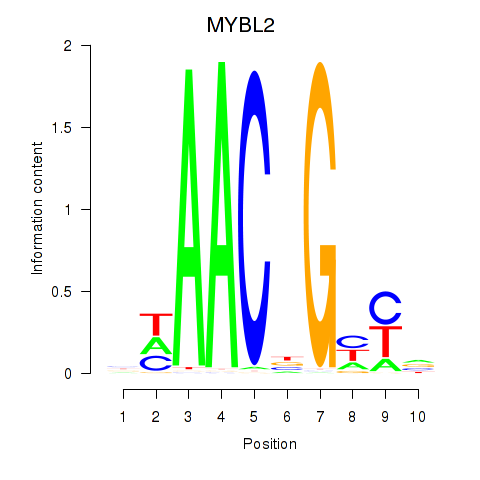
Results for MYBL2
Z-value: 1.34
Transcription factors associated with MYBL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL2
|
ENSG00000101057.11 | MYBL2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL2 | hg19_v2_chr20_+_42295745_42295797 | 0.65 | 7.0e-03 | Click! |
Activity profile of MYBL2 motif
Sorted Z-values of MYBL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_14530143 | 1.85 |
ENST00000242776.4 |
DDX39A |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr2_+_127413481 | 1.80 |
ENST00000259254.4 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr11_+_62104897 | 1.79 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr3_+_157154578 | 1.69 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_+_91966384 | 1.69 |
ENST00000430031.2 ENST00000234626.6 |
CDC7 |
cell division cycle 7 |
| chrX_+_52780318 | 1.66 |
ENST00000375515.3 ENST00000276049.6 |
SSX2B |
synovial sarcoma, X breakpoint 2B |
| chr10_+_31608054 | 1.62 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr17_+_45286387 | 1.49 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_+_127413677 | 1.43 |
ENST00000356887.7 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr5_-_94417339 | 1.31 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr1_+_91966656 | 1.28 |
ENST00000428239.1 ENST00000426137.1 |
CDC7 |
cell division cycle 7 |
| chr12_+_93965609 | 1.22 |
ENST00000549887.1 ENST00000551556.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr11_-_9025541 | 1.21 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr1_+_45205478 | 1.21 |
ENST00000452259.1 ENST00000372224.4 |
KIF2C |
kinesin family member 2C |
| chr1_+_45205498 | 1.21 |
ENST00000372218.4 |
KIF2C |
kinesin family member 2C |
| chr12_+_93965451 | 1.20 |
ENST00000548537.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr17_+_45286706 | 1.12 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr15_-_55611306 | 1.12 |
ENST00000563262.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_+_93771659 | 1.10 |
ENST00000337179.5 ENST00000415493.2 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr1_-_114301503 | 1.09 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chrX_+_65384182 | 1.06 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chrX_+_65384052 | 1.04 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr12_+_93964746 | 1.04 |
ENST00000536696.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chrX_-_48056199 | 1.02 |
ENST00000311798.1 ENST00000347757.1 |
SSX5 |
synovial sarcoma, X breakpoint 5 |
| chr1_-_85156216 | 1.01 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr6_+_27806319 | 0.99 |
ENST00000606613.1 ENST00000396980.3 |
HIST1H2BN |
histone cluster 1, H2bn |
| chr4_+_41258786 | 0.95 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chrX_+_48242863 | 0.90 |
ENST00000376886.2 ENST00000375517.3 |
SSX4 |
synovial sarcoma, X breakpoint 4 |
| chr4_-_16900410 | 0.90 |
ENST00000304523.5 |
LDB2 |
LIM domain binding 2 |
| chr19_-_55919087 | 0.90 |
ENST00000587845.1 ENST00000589978.1 ENST00000264552.9 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
| chr5_-_88179302 | 0.90 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr19_-_39826639 | 0.90 |
ENST00000602185.1 ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG |
glia maturation factor, gamma |
| chr19_-_8373173 | 0.89 |
ENST00000537716.2 ENST00000301458.5 |
CD320 |
CD320 molecule |
| chr15_-_91537723 | 0.89 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr4_-_16900242 | 0.89 |
ENST00000502640.1 ENST00000506732.1 |
LDB2 |
LIM domain binding 2 |
| chr12_-_110906027 | 0.88 |
ENST00000537466.2 ENST00000550974.1 ENST00000228827.3 |
GPN3 |
GPN-loop GTPase 3 |
| chr1_+_211499957 | 0.87 |
ENST00000336184.2 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr2_+_47630255 | 0.85 |
ENST00000406134.1 |
MSH2 |
mutS homolog 2 |
| chr1_+_246887349 | 0.85 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr1_+_211500129 | 0.84 |
ENST00000427925.2 ENST00000261464.5 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr10_+_94352956 | 0.83 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr18_+_32558208 | 0.82 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_23146271 | 0.80 |
ENST00000545771.1 |
KLHL7 |
kelch-like family member 7 |
| chr1_-_114301960 | 0.80 |
ENST00000369598.1 ENST00000369600.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr1_-_163172625 | 0.80 |
ENST00000527988.1 ENST00000531476.1 ENST00000530507.1 |
RGS5 |
regulator of G-protein signaling 5 |
| chr2_+_47630108 | 0.79 |
ENST00000233146.2 ENST00000454849.1 ENST00000543555.1 |
MSH2 |
mutS homolog 2 |
| chr12_-_71031185 | 0.79 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr5_-_88179017 | 0.78 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chrX_-_48216101 | 0.78 |
ENST00000298396.2 ENST00000376893.3 |
SSX3 |
synovial sarcoma, X breakpoint 3 |
| chr17_+_34948228 | 0.77 |
ENST00000251312.5 ENST00000590554.1 |
DHRS11 |
dehydrogenase/reductase (SDR family) member 11 |
| chrX_-_48271344 | 0.76 |
ENST00000376884.2 ENST00000396928.1 |
SSX4B |
synovial sarcoma, X breakpoint 4B |
| chr14_-_22005018 | 0.75 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr1_-_114301755 | 0.74 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr17_+_7155819 | 0.74 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr12_+_110906169 | 0.73 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr11_+_128563652 | 0.73 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_46049706 | 0.72 |
ENST00000527470.1 ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
| chr11_-_28129656 | 0.71 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr3_+_173116225 | 0.71 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr4_+_40192656 | 0.70 |
ENST00000505618.1 |
RHOH |
ras homolog family member H |
| chr17_-_38978847 | 0.69 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr17_+_7155556 | 0.69 |
ENST00000570500.1 ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr7_+_116654935 | 0.69 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr17_+_76210367 | 0.69 |
ENST00000592734.1 ENST00000587746.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr16_+_66586461 | 0.69 |
ENST00000264001.4 ENST00000351137.4 ENST00000345436.4 ENST00000362093.4 ENST00000417030.2 ENST00000527729.1 ENST00000532838.1 |
CKLF CKLF-CMTM1 |
chemokine-like factor CKLF-CMTM1 readthrough |
| chr11_+_34073757 | 0.68 |
ENST00000532820.1 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr6_-_13711773 | 0.67 |
ENST00000011619.3 |
RANBP9 |
RAN binding protein 9 |
| chr14_-_22005343 | 0.66 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chr10_+_35415719 | 0.66 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr14_-_22005062 | 0.65 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chr11_-_32452357 | 0.64 |
ENST00000379079.2 ENST00000530998.1 |
WT1 |
Wilms tumor 1 |
| chr7_+_98476095 | 0.64 |
ENST00000359863.4 ENST00000355540.3 |
TRRAP |
transformation/transcription domain-associated protein |
| chr4_-_77069533 | 0.64 |
ENST00000514987.1 ENST00000458189.2 ENST00000514901.1 ENST00000342467.6 |
NUP54 |
nucleoporin 54kDa |
| chr17_-_38574169 | 0.64 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr1_-_114302086 | 0.63 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr2_+_48010221 | 0.63 |
ENST00000234420.5 |
MSH6 |
mutS homolog 6 |
| chr1_+_196788887 | 0.63 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr3_+_150126101 | 0.62 |
ENST00000361875.3 ENST00000361136.2 |
TSC22D2 |
TSC22 domain family, member 2 |
| chr12_+_123011776 | 0.62 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr19_-_12912657 | 0.60 |
ENST00000301522.2 |
PRDX2 |
peroxiredoxin 2 |
| chr16_+_20817761 | 0.59 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr4_-_77069573 | 0.58 |
ENST00000264883.3 |
NUP54 |
nucleoporin 54kDa |
| chr7_-_127983877 | 0.58 |
ENST00000415472.2 ENST00000478061.1 ENST00000223073.2 ENST00000459726.1 |
RBM28 |
RNA binding motif protein 28 |
| chr16_-_3285144 | 0.58 |
ENST00000431561.3 ENST00000396870.4 |
ZNF200 |
zinc finger protein 200 |
| chr19_-_12912601 | 0.58 |
ENST00000334482.5 |
PRDX2 |
peroxiredoxin 2 |
| chr16_-_31076332 | 0.58 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr22_-_42342692 | 0.57 |
ENST00000404067.1 ENST00000402338.1 |
CENPM |
centromere protein M |
| chr4_+_17812525 | 0.56 |
ENST00000251496.2 |
NCAPG |
non-SMC condensin I complex, subunit G |
| chr17_+_7155343 | 0.56 |
ENST00000573513.1 ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chrX_-_71458802 | 0.56 |
ENST00000373657.1 ENST00000334463.3 |
ERCC6L |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr16_+_20817839 | 0.56 |
ENST00000348433.6 ENST00000568501.1 ENST00000566276.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr5_-_137667459 | 0.56 |
ENST00000415130.2 ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C |
cell division cycle 25C |
| chr1_-_26232522 | 0.55 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr1_-_109935819 | 0.55 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr7_-_128694927 | 0.54 |
ENST00000471166.1 ENST00000265388.5 |
TNPO3 |
transportin 3 |
| chr8_+_37963311 | 0.54 |
ENST00000428278.2 ENST00000521652.1 |
ASH2L |
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr17_+_76210267 | 0.54 |
ENST00000301633.4 ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr10_+_35415978 | 0.54 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr6_+_34725181 | 0.53 |
ENST00000244520.5 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr16_+_29802036 | 0.52 |
ENST00000561482.1 ENST00000160827.4 ENST00000569636.2 ENST00000400750.2 |
KIF22 |
kinesin family member 22 |
| chr3_-_182817297 | 0.52 |
ENST00000539926.1 ENST00000476176.1 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr21_-_46237883 | 0.52 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr13_-_76056250 | 0.51 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr5_-_137667526 | 0.50 |
ENST00000503022.1 |
CDC25C |
cell division cycle 25C |
| chr1_-_12677714 | 0.50 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr16_+_20818020 | 0.50 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr7_+_23145884 | 0.50 |
ENST00000409689.1 ENST00000410047.1 |
KLHL7 |
kelch-like family member 7 |
| chr3_-_3221358 | 0.50 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr7_-_148581360 | 0.49 |
ENST00000320356.2 ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr1_+_212208919 | 0.49 |
ENST00000366991.4 ENST00000542077.1 |
DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr14_+_54863739 | 0.49 |
ENST00000541304.1 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr15_+_81475047 | 0.49 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr3_-_88108212 | 0.48 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr5_-_137674000 | 0.48 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr10_-_53459319 | 0.48 |
ENST00000331173.4 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr8_+_6565854 | 0.47 |
ENST00000285518.6 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr1_-_39339777 | 0.47 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chrX_-_153775426 | 0.47 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr3_-_98241358 | 0.46 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr8_-_28747424 | 0.46 |
ENST00000523436.1 ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9 |
integrator complex subunit 9 |
| chr5_+_37379314 | 0.46 |
ENST00000265107.4 ENST00000504564.1 |
WDR70 |
WD repeat domain 70 |
| chr7_+_73097890 | 0.45 |
ENST00000265758.2 ENST00000423166.2 ENST00000423497.1 |
WBSCR22 |
Williams Beuren syndrome chromosome region 22 |
| chr22_+_20067738 | 0.45 |
ENST00000351989.3 ENST00000383024.2 |
DGCR8 |
DGCR8 microprocessor complex subunit |
| chr1_-_171711177 | 0.45 |
ENST00000415773.1 ENST00000367740.2 |
VAMP4 |
vesicle-associated membrane protein 4 |
| chr6_-_32098013 | 0.45 |
ENST00000375156.3 |
FKBPL |
FK506 binding protein like |
| chr7_-_16505440 | 0.45 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chrX_+_129473916 | 0.45 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr4_+_110736659 | 0.44 |
ENST00000394631.3 ENST00000226796.6 |
GAR1 |
GAR1 ribonucleoprotein |
| chr3_-_186285077 | 0.43 |
ENST00000338733.5 |
TBCCD1 |
TBCC domain containing 1 |
| chr12_-_6961050 | 0.43 |
ENST00000538862.2 |
CDCA3 |
cell division cycle associated 3 |
| chr11_+_34073872 | 0.43 |
ENST00000530820.1 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr14_+_54863667 | 0.43 |
ENST00000335183.6 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr17_+_61678225 | 0.43 |
ENST00000258975.6 |
TACO1 |
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr1_+_66258846 | 0.43 |
ENST00000341517.4 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr3_-_185655795 | 0.43 |
ENST00000342294.4 ENST00000382191.4 ENST00000453386.2 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
| chr12_+_12870055 | 0.43 |
ENST00000228872.4 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr12_-_104532062 | 0.42 |
ENST00000240055.3 |
NFYB |
nuclear transcription factor Y, beta |
| chr14_+_54863682 | 0.42 |
ENST00000543789.2 ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr1_+_24286287 | 0.42 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr12_-_71031220 | 0.41 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr6_-_43595039 | 0.40 |
ENST00000307114.7 |
GTPBP2 |
GTP binding protein 2 |
| chr3_+_160939050 | 0.40 |
ENST00000493066.1 ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3 |
NMD3 ribosome export adaptor |
| chr7_+_138145076 | 0.40 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr7_+_100464760 | 0.40 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr3_-_165555200 | 0.40 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr6_+_36562132 | 0.40 |
ENST00000373715.6 ENST00000339436.7 |
SRSF3 |
serine/arginine-rich splicing factor 3 |
| chr13_-_23949671 | 0.39 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr9_+_116267536 | 0.39 |
ENST00000374136.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr4_+_54243862 | 0.39 |
ENST00000306932.6 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr16_+_85646763 | 0.39 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr9_-_130637244 | 0.39 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr1_+_163038565 | 0.39 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr7_+_99006232 | 0.39 |
ENST00000403633.2 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr1_-_197115818 | 0.39 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr20_+_43160458 | 0.39 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr12_-_68726052 | 0.38 |
ENST00000540418.1 ENST00000411698.2 ENST00000393543.3 ENST00000303145.7 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr2_+_48010312 | 0.38 |
ENST00000540021.1 |
MSH6 |
mutS homolog 6 |
| chr4_+_128802016 | 0.38 |
ENST00000270861.5 ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4 |
polo-like kinase 4 |
| chr16_-_4896205 | 0.38 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr11_-_10829851 | 0.37 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr10_+_71211212 | 0.37 |
ENST00000373290.2 |
TSPAN15 |
tetraspanin 15 |
| chr15_+_75182346 | 0.37 |
ENST00000569931.1 ENST00000352410.4 ENST00000566377.1 ENST00000569233.1 ENST00000567132.1 ENST00000564633.1 ENST00000568907.1 ENST00000563422.1 ENST00000564003.1 ENST00000562800.1 ENST00000563786.1 ENST00000535694.1 ENST00000323744.6 ENST00000568828.1 ENST00000562606.1 ENST00000565576.1 ENST00000567570.1 |
MPI |
mannose phosphate isomerase |
| chr19_+_49496705 | 0.37 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr1_+_174843548 | 0.36 |
ENST00000478442.1 ENST00000465412.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr16_-_57481278 | 0.36 |
ENST00000567751.1 ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1 |
cytokine induced apoptosis inhibitor 1 |
| chr4_+_54243798 | 0.36 |
ENST00000337488.6 ENST00000358575.5 ENST00000507922.1 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr22_-_42343117 | 0.36 |
ENST00000407253.3 ENST00000215980.5 |
CENPM |
centromere protein M |
| chr3_+_49027308 | 0.36 |
ENST00000383729.4 ENST00000343546.4 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr2_+_143886877 | 0.35 |
ENST00000295095.6 |
ARHGAP15 |
Rho GTPase activating protein 15 |
| chr1_-_155990580 | 0.35 |
ENST00000531917.1 ENST00000480567.1 ENST00000526212.1 ENST00000529008.1 ENST00000496742.1 ENST00000295702.4 |
SSR2 |
signal sequence receptor, beta (translocon-associated protein beta) |
| chr3_-_88108192 | 0.35 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr6_+_170863421 | 0.35 |
ENST00000392092.2 ENST00000540980.1 ENST00000230354.6 |
TBP |
TATA box binding protein |
| chr1_+_32666188 | 0.35 |
ENST00000421922.2 |
CCDC28B |
coiled-coil domain containing 28B |
| chr17_-_66951474 | 0.34 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr8_-_121457332 | 0.34 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr2_-_128784846 | 0.34 |
ENST00000259235.3 ENST00000357702.5 ENST00000424298.1 |
SAP130 |
Sin3A-associated protein, 130kDa |
| chr2_-_70475730 | 0.34 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_-_29233769 | 0.34 |
ENST00000581216.1 |
TEFM |
transcription elongation factor, mitochondrial |
| chr2_-_70475701 | 0.34 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr8_-_121457608 | 0.34 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr1_-_114355083 | 0.34 |
ENST00000261441.5 |
RSBN1 |
round spermatid basic protein 1 |
| chr3_+_99357319 | 0.34 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr6_+_150070857 | 0.34 |
ENST00000544496.1 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_-_173176452 | 0.33 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr5_-_43557791 | 0.33 |
ENST00000338972.4 ENST00000511321.1 ENST00000515338.1 |
PAIP1 |
poly(A) binding protein interacting protein 1 |
| chr20_+_55926583 | 0.33 |
ENST00000395840.2 |
RAE1 |
ribonucleic acid export 1 |
| chr17_-_36413133 | 0.33 |
ENST00000523089.1 ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2 |
TBC1 domain family member 3 |
| chr21_-_35014027 | 0.33 |
ENST00000399442.1 ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr8_-_67976509 | 0.33 |
ENST00000518747.1 |
COPS5 |
COP9 signalosome subunit 5 |
| chr11_+_34073269 | 0.33 |
ENST00000389645.3 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr10_-_74385811 | 0.33 |
ENST00000603011.1 ENST00000401998.3 ENST00000361114.5 ENST00000604238.1 |
MICU1 |
mitochondrial calcium uptake 1 |
| chr1_-_243418621 | 0.32 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr7_-_100076873 | 0.32 |
ENST00000300181.2 |
TSC22D4 |
TSC22 domain family, member 4 |
| chrX_+_129473859 | 0.32 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_-_109825719 | 0.32 |
ENST00000369904.3 ENST00000369903.2 ENST00000429031.1 ENST00000418914.2 ENST00000409267.1 |
PSRC1 |
proline/serine-rich coiled-coil 1 |
| chr14_-_75530693 | 0.32 |
ENST00000555135.1 ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr17_+_21730180 | 0.32 |
ENST00000584398.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr19_+_50528971 | 0.32 |
ENST00000598809.1 ENST00000595661.1 ENST00000391821.2 |
ZNF473 |
zinc finger protein 473 |
| chr8_-_94753229 | 0.32 |
ENST00000518597.1 ENST00000399300.2 ENST00000517700.1 |
RBM12B |
RNA binding motif protein 12B |
| chr3_+_9834179 | 0.32 |
ENST00000498623.2 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 2.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 1.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.4 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.5 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 2.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.4 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 2.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 3.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.4 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 2.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 4.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.7 | 2.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.6 | 1.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.6 | 3.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 2.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 0.9 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.2 | 0.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 2.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.1 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.8 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.5 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.3 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.5 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 0.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.0 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0034594 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 3.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 1.7 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 1.6 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.3 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 1.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.3 | 0.9 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 2.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.7 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 1.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 3.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 1.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 0.5 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 0.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.6 | GO:0072299 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 1.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.4 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.1 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.5 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 0.1 | 0.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.3 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 2.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.1 | 1.8 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:1900195 | spindle assembly involved in female meiosis I(GO:0007057) positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.4 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 3.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.3 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 2.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.3 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.9 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0035607 | vacuolar phosphate transport(GO:0007037) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.0 | 1.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 2.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 3.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0002436 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.0 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.1 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.1 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 1.2 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0060702 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0019050 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.0 | 0.7 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.4 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 2.1 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 3.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |