Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
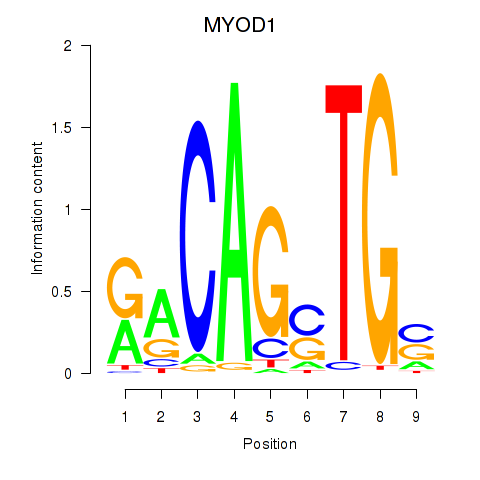
Results for MYOD1
Z-value: 1.98
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | MYOD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg19_v2_chr11_+_17741111_17741124 | 0.93 | 1.1e-07 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175629135 | 9.24 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_-_201346761 | 6.65 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr16_+_30387141 | 6.57 |
ENST00000566955.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_-_152590946 | 5.72 |
ENST00000172853.10 |
NEB |
nebulin |
| chr16_+_30383613 | 5.58 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr16_+_30386098 | 5.11 |
ENST00000322861.7 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_-_175629164 | 4.86 |
ENST00000409323.1 ENST00000261007.5 ENST00000348749.5 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_-_201391149 | 4.73 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr9_-_35689900 | 4.67 |
ENST00000378300.5 ENST00000329305.2 ENST00000360958.2 |
TPM2 |
tropomyosin 2 (beta) |
| chr15_-_35088340 | 4.60 |
ENST00000290378.4 |
ACTC1 |
actin, alpha, cardiac muscle 1 |
| chr2_+_170366203 | 4.59 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr1_-_193155729 | 4.51 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_-_35685452 | 4.26 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chrX_-_107018969 | 4.05 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr3_-_52486841 | 3.97 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr21_+_30502806 | 3.92 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr17_+_45286706 | 3.90 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_+_220283091 | 3.84 |
ENST00000373960.3 |
DES |
desmin |
| chr17_+_45286387 | 3.76 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_-_47470591 | 3.58 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr6_+_44184653 | 3.53 |
ENST00000573382.2 ENST00000576476.1 |
RP1-302G2.5 |
RP1-302G2.5 |
| chr3_+_35721106 | 3.47 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr9_-_79307096 | 3.38 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr11_-_47470703 | 3.29 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr10_+_11060004 | 3.22 |
ENST00000542579.1 ENST00000399850.3 ENST00000417956.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr2_-_152590982 | 3.21 |
ENST00000409198.1 ENST00000397345.3 ENST00000427231.2 |
NEB |
nebulin |
| chr11_-_2170786 | 3.05 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr11_-_47470682 | 2.93 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr4_-_186456652 | 2.89 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_186456766 | 2.88 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr15_-_48937982 | 2.88 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr10_-_75410771 | 2.64 |
ENST00000372873.4 |
SYNPO2L |
synaptopodin 2-like |
| chr21_+_27011899 | 2.36 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr2_-_37899323 | 2.34 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_-_179672142 | 2.28 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr4_-_157892498 | 2.28 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr17_+_48133459 | 2.25 |
ENST00000320031.8 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr21_+_27011584 | 2.18 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr8_-_41522779 | 2.12 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr19_-_45826125 | 2.02 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr11_-_111783595 | 1.97 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr11_+_86511569 | 1.91 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr2_-_175870085 | 1.89 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr13_+_102104980 | 1.84 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_+_116165754 | 1.84 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr13_+_102104952 | 1.79 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_+_169418195 | 1.73 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr20_-_44455976 | 1.71 |
ENST00000372555.3 |
TNNC2 |
troponin C type 2 (fast) |
| chrX_-_107019181 | 1.71 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr16_+_6533380 | 1.70 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_41522719 | 1.70 |
ENST00000335651.6 |
ANK1 |
ankyrin 1, erythrocytic |
| chr12_-_50677255 | 1.70 |
ENST00000551691.1 ENST00000394943.3 ENST00000341247.4 |
LIMA1 |
LIM domain and actin binding 1 |
| chr16_+_7382745 | 1.69 |
ENST00000436368.2 ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_+_172422026 | 1.68 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr11_+_44587141 | 1.67 |
ENST00000227155.4 ENST00000342935.3 ENST00000532544.1 |
CD82 |
CD82 molecule |
| chr3_+_159570722 | 1.65 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr3_+_35681081 | 1.65 |
ENST00000428373.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chrX_+_70521584 | 1.63 |
ENST00000373829.3 ENST00000538820.1 |
ITGB1BP2 |
integrin beta 1 binding protein (melusin) 2 |
| chrX_-_6146876 | 1.57 |
ENST00000381095.3 |
NLGN4X |
neuroligin 4, X-linked |
| chr7_+_73442487 | 1.53 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chrX_-_13835147 | 1.52 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr17_+_52978156 | 1.51 |
ENST00000348161.4 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chr4_+_169418255 | 1.50 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr13_+_76334795 | 1.49 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr7_+_73442422 | 1.48 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr13_+_76334567 | 1.47 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr10_+_11059826 | 1.47 |
ENST00000450189.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr12_+_79258444 | 1.44 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr7_-_151574191 | 1.40 |
ENST00000287878.4 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_+_52978107 | 1.38 |
ENST00000445275.2 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chr17_+_65040678 | 1.37 |
ENST00000226021.3 |
CACNG1 |
calcium channel, voltage-dependent, gamma subunit 1 |
| chr4_-_41216473 | 1.36 |
ENST00000513140.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr15_-_42749711 | 1.36 |
ENST00000565611.1 ENST00000263805.4 ENST00000565948.1 |
ZNF106 |
zinc finger protein 106 |
| chr3_-_178790057 | 1.35 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr6_-_24911195 | 1.34 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr1_-_86043921 | 1.32 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr12_+_79258547 | 1.27 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr1_+_25071848 | 1.23 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr4_-_41216492 | 1.22 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr9_+_90112767 | 1.21 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr11_+_1860200 | 1.21 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr2_-_197041193 | 1.20 |
ENST00000409228.1 |
STK17B |
serine/threonine kinase 17b |
| chr11_+_1942580 | 1.19 |
ENST00000381558.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr10_+_96162242 | 1.19 |
ENST00000225235.4 |
TBC1D12 |
TBC1 domain family, member 12 |
| chr15_+_32933866 | 1.16 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr3_-_87040233 | 1.14 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr6_-_31697255 | 1.14 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_73442457 | 1.12 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr5_-_150466692 | 1.08 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr1_-_161993422 | 1.07 |
ENST00000367940.2 |
OLFML2B |
olfactomedin-like 2B |
| chr6_+_41021027 | 1.07 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr1_-_94312706 | 1.06 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr4_-_41216619 | 1.04 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr14_+_90863327 | 1.03 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr16_-_67427389 | 1.02 |
ENST00000562206.1 ENST00000290942.5 ENST00000393957.2 |
TPPP3 |
tubulin polymerization-promoting protein family member 3 |
| chr11_+_1860832 | 1.01 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr5_+_49962495 | 1.00 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr7_+_73442102 | 1.00 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chr7_-_558876 | 0.99 |
ENST00000354513.5 ENST00000402802.3 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
| chr5_+_34656569 | 0.99 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr19_-_49658387 | 0.99 |
ENST00000595625.1 |
HRC |
histidine rich calcium binding protein |
| chr6_-_137113604 | 0.96 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chrX_-_106243451 | 0.95 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr4_-_164534657 | 0.95 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr11_-_73694346 | 0.95 |
ENST00000310473.3 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_1860682 | 0.95 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr3_-_178789993 | 0.92 |
ENST00000432729.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr3_+_30647994 | 0.91 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr1_-_17304771 | 0.89 |
ENST00000375534.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr15_-_45670924 | 0.88 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chrX_-_153637612 | 0.88 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr3_+_30648066 | 0.88 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr10_-_118032979 | 0.88 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr8_-_29120580 | 0.87 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr2_+_24714729 | 0.87 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr10_-_118032697 | 0.87 |
ENST00000439649.3 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr22_+_38071615 | 0.87 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr4_+_47487285 | 0.87 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr19_+_3933579 | 0.86 |
ENST00000593949.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr11_-_6341844 | 0.85 |
ENST00000303927.3 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr1_-_33168336 | 0.81 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr12_+_122150646 | 0.80 |
ENST00000449592.2 |
TMEM120B |
transmembrane protein 120B |
| chr11_+_1940786 | 0.78 |
ENST00000278317.6 ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr11_-_64013288 | 0.77 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_64014379 | 0.77 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_-_154164534 | 0.74 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr1_-_154928562 | 0.71 |
ENST00000368463.3 ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
| chr9_-_16727978 | 0.68 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr1_-_16345245 | 0.68 |
ENST00000311890.9 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr11_-_119252359 | 0.68 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr1_-_204329013 | 0.68 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr19_-_49658641 | 0.63 |
ENST00000252825.4 |
HRC |
histidine rich calcium binding protein |
| chr15_-_72523454 | 0.62 |
ENST00000565154.1 ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM |
pyruvate kinase, muscle |
| chrX_-_13835461 | 0.62 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr1_-_40367668 | 0.61 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr9_+_4679555 | 0.60 |
ENST00000381858.1 ENST00000381854.3 |
CDC37L1 |
cell division cycle 37-like 1 |
| chr7_-_92465868 | 0.60 |
ENST00000424848.2 |
CDK6 |
cyclin-dependent kinase 6 |
| chr10_+_123923105 | 0.60 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_119252425 | 0.59 |
ENST00000260187.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr18_-_33702078 | 0.59 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr7_+_18535786 | 0.59 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr12_-_110434021 | 0.59 |
ENST00000355312.3 ENST00000551209.1 ENST00000550186.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr3_-_57113314 | 0.59 |
ENST00000338458.4 ENST00000468727.1 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr6_-_3227877 | 0.59 |
ENST00000259818.7 |
TUBB2B |
tubulin, beta 2B class IIb |
| chr12_-_92539614 | 0.58 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr19_+_3933085 | 0.58 |
ENST00000168977.2 ENST00000599576.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr9_-_13165457 | 0.58 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr4_-_52904425 | 0.58 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr3_+_99357319 | 0.57 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr2_+_121103706 | 0.56 |
ENST00000295228.3 |
INHBB |
inhibin, beta B |
| chr8_-_101963482 | 0.55 |
ENST00000419477.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_+_43299241 | 0.55 |
ENST00000328118.3 |
FMNL1 |
formin-like 1 |
| chr12_+_56473628 | 0.55 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr16_-_31076273 | 0.54 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr3_+_46919235 | 0.54 |
ENST00000449590.1 |
PTH1R |
parathyroid hormone 1 receptor |
| chr8_-_101962777 | 0.54 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_+_123922941 | 0.51 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr11_+_33279850 | 0.51 |
ENST00000531504.1 ENST00000456517.1 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr11_-_67120974 | 0.50 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr17_+_66508537 | 0.49 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_+_148402069 | 0.49 |
ENST00000358556.4 ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA |
endothelin receptor type A |
| chr10_+_123923205 | 0.49 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr21_+_38071430 | 0.48 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr16_+_31044812 | 0.48 |
ENST00000313843.3 |
STX4 |
syntaxin 4 |
| chr11_+_71938925 | 0.48 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr11_-_6341724 | 0.46 |
ENST00000530979.1 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr16_-_31076332 | 0.46 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr2_-_220435963 | 0.45 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr17_+_43299156 | 0.45 |
ENST00000331495.3 |
FMNL1 |
formin-like 1 |
| chr19_+_48216600 | 0.45 |
ENST00000263277.3 ENST00000538399.1 |
EHD2 |
EH-domain containing 2 |
| chr15_-_42186248 | 0.45 |
ENST00000320955.6 |
SPTBN5 |
spectrin, beta, non-erythrocytic 5 |
| chr11_-_10829851 | 0.45 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr19_+_16187085 | 0.44 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr16_-_122619 | 0.43 |
ENST00000262316.6 |
RHBDF1 |
rhomboid 5 homolog 1 (Drosophila) |
| chr17_+_66508154 | 0.43 |
ENST00000358598.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr7_+_107301065 | 0.43 |
ENST00000265715.3 |
SLC26A4 |
solute carrier family 26 (anion exchanger), member 4 |
| chr1_-_84464780 | 0.42 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr18_-_53089723 | 0.42 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr12_-_56122426 | 0.42 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr1_+_156105878 | 0.41 |
ENST00000508500.1 |
LMNA |
lamin A/C |
| chr12_-_56122761 | 0.40 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr15_-_30113676 | 0.40 |
ENST00000400011.2 |
TJP1 |
tight junction protein 1 |
| chr12_+_14518598 | 0.40 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr16_+_31044413 | 0.40 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr18_-_52989525 | 0.39 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr5_-_148758839 | 0.39 |
ENST00000261796.3 |
IL17B |
interleukin 17B |
| chr3_-_141868293 | 0.38 |
ENST00000317104.7 ENST00000494358.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_81046841 | 0.38 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr5_+_131630117 | 0.38 |
ENST00000200652.3 |
SLC22A4 |
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
| chr8_-_145018080 | 0.37 |
ENST00000354589.3 |
PLEC |
plectin |
| chr1_+_87170247 | 0.37 |
ENST00000370558.4 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
| chr17_+_68165657 | 0.36 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr3_-_99833333 | 0.36 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr16_+_57673430 | 0.36 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_+_87170577 | 0.36 |
ENST00000482504.1 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
| chr2_-_128400788 | 0.35 |
ENST00000409286.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr5_-_58335281 | 0.35 |
ENST00000358923.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr1_+_89990431 | 0.35 |
ENST00000330947.2 ENST00000358200.4 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
| chr13_-_99630233 | 0.34 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr8_-_130951940 | 0.34 |
ENST00000522250.1 ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B |
family with sequence similarity 49, member B |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 16.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.0 | 5.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.8 | 9.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.7 | 2.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.6 | 11.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 13.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.6 | 20.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.5 | 2.7 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 10.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.5 | 5.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 5.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 14.8 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 1.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 3.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 23.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.0 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 2.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 3.7 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 3.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.7 | 50.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 4.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 3.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 2.7 | 10.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.2 | 8.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 1.8 | 14.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 1.0 | 6.8 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.9 | 2.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.7 | 5.8 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.6 | 1.8 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.6 | 1.8 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.5 | 1.6 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.5 | 2.6 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.5 | 4.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 6.9 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.5 | 33.3 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.4 | 0.9 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.4 | 1.1 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 3.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 0.9 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.3 | 0.9 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 2.9 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 1.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 0.2 | GO:0071415 | response to caffeine(GO:0031000) cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 4.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 2.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.6 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.2 | 0.6 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 2.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 0.5 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.2 | 2.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.7 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 3.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 1.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 4.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.6 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 2.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 2.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.0 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 1.8 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.1 | 15.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 0.4 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.3 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.1 | 1.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 5.0 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.8 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 3.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.6 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.1 | GO:0061299 | extracellular matrix-cell signaling(GO:0035426) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 1.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.6 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 3.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 2.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 2.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.9 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.4 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 3.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.9 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 2.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.0 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.5 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.3 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 1.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0071896 | negative regulation of hippo signaling(GO:0035331) protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.5 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 3.8 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 1.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 1.5 | 4.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.1 | 12.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.6 | 5.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.6 | 1.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.6 | 13.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.5 | 9.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.5 | 2.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 2.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 45.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 1.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 1.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 2.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 3.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 3.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 0.9 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 3.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 0.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 3.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 4.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 4.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 3.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 4.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0034594 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 2.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 3.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 3.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 4.7 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 4.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) AMP binding(GO:0016208) |
| 0.0 | 2.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |