Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
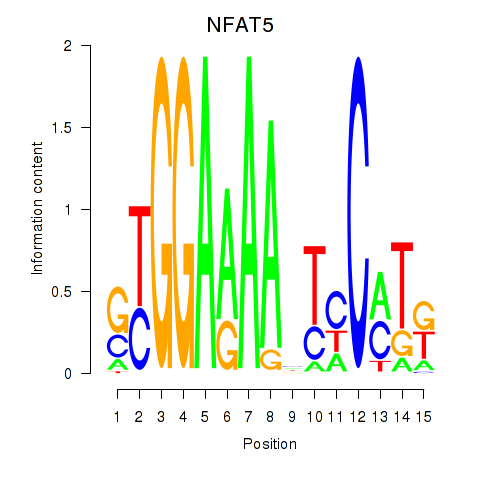
Results for NFAT5
Z-value: 1.03
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.16 | NFAT5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFAT5 | hg19_v2_chr16_+_69600209_69600254 | 0.15 | 5.7e-01 | Click! |
Activity profile of NFAT5 motif
Sorted Z-values of NFAT5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFAT5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_47579188 | 3.35 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr1_-_153517473 | 3.11 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr1_-_57045228 | 2.76 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr6_+_86159821 | 2.66 |
ENST00000369651.3 |
NT5E |
5'-nucleotidase, ecto (CD73) |
| chr12_+_26274917 | 2.65 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr11_-_111794446 | 2.15 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr14_-_25479811 | 2.10 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr14_-_30396948 | 1.97 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr11_+_12399071 | 1.51 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr3_+_8543393 | 1.51 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr14_-_102706934 | 1.49 |
ENST00000523231.1 ENST00000524370.1 ENST00000517966.1 |
MOK |
MOK protein kinase |
| chr3_+_8543561 | 1.44 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr13_-_67804445 | 1.43 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr1_-_201346761 | 1.41 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr11_+_19799327 | 1.40 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr8_-_49833978 | 1.39 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr10_+_89419370 | 1.35 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr11_+_19798964 | 1.35 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr11_+_131240373 | 1.29 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr5_-_150521192 | 1.19 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr7_+_128399002 | 1.12 |
ENST00000493278.1 |
CALU |
calumenin |
| chr2_-_162931052 | 1.11 |
ENST00000360534.3 |
DPP4 |
dipeptidyl-peptidase 4 |
| chr3_+_8543533 | 1.03 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr18_-_33702078 | 1.02 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr12_+_10365404 | 0.98 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr11_+_131781290 | 0.94 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr7_+_65670186 | 0.83 |
ENST00000304842.5 ENST00000442120.1 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
| chr8_-_27469196 | 0.72 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr4_-_76861392 | 0.70 |
ENST00000505594.1 |
NAAA |
N-acylethanolamine acid amidase |
| chr3_-_47950745 | 0.66 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr6_-_131291572 | 0.63 |
ENST00000529208.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr9_-_13175823 | 0.63 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr4_-_74847800 | 0.62 |
ENST00000296029.3 |
PF4 |
platelet factor 4 |
| chr4_+_71587669 | 0.62 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr3_+_49027308 | 0.58 |
ENST00000383729.4 ENST00000343546.4 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr6_+_89790459 | 0.57 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chrX_+_37208540 | 0.57 |
ENST00000466533.1 ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1 TM4SF2 |
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr19_+_17420340 | 0.55 |
ENST00000359866.4 |
DDA1 |
DET1 and DDB1 associated 1 |
| chr4_+_26322185 | 0.55 |
ENST00000361572.6 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_115300579 | 0.54 |
ENST00000358528.4 ENST00000525132.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr16_+_89988259 | 0.53 |
ENST00000554444.1 ENST00000556565.1 |
TUBB3 |
Tubulin beta-3 chain |
| chr6_-_134495992 | 0.52 |
ENST00000475719.2 ENST00000367857.5 ENST00000237305.7 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chrX_+_37208521 | 0.50 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr12_-_123756781 | 0.49 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr1_-_115300592 | 0.48 |
ENST00000261443.5 ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr4_+_26322409 | 0.48 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_16345245 | 0.48 |
ENST00000311890.9 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr12_+_69139886 | 0.44 |
ENST00000398004.2 |
SLC35E3 |
solute carrier family 35, member E3 |
| chr6_+_89790490 | 0.44 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr11_-_67120974 | 0.43 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr15_+_49170083 | 0.43 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr19_+_48828582 | 0.40 |
ENST00000270221.6 ENST00000596315.1 |
EMP3 |
epithelial membrane protein 3 |
| chr19_-_46285736 | 0.38 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr5_-_77844974 | 0.35 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr20_+_33104199 | 0.33 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr3_-_149293990 | 0.33 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr14_+_100848311 | 0.32 |
ENST00000542471.2 |
WDR25 |
WD repeat domain 25 |
| chr7_-_100888337 | 0.29 |
ENST00000223136.4 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr13_-_24463530 | 0.28 |
ENST00000382172.3 |
MIPEP |
mitochondrial intermediate peptidase |
| chr14_+_73704201 | 0.28 |
ENST00000340738.5 ENST00000427855.1 ENST00000381166.3 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr19_+_48828788 | 0.27 |
ENST00000594198.1 ENST00000597279.1 ENST00000593437.1 |
EMP3 |
epithelial membrane protein 3 |
| chrX_-_41782592 | 0.27 |
ENST00000378158.1 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr11_+_8932828 | 0.26 |
ENST00000530281.1 ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr1_+_18958008 | 0.26 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr20_-_23731893 | 0.26 |
ENST00000398402.1 |
CST1 |
cystatin SN |
| chr17_+_18684563 | 0.26 |
ENST00000476139.1 |
TVP23B |
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr1_-_243418344 | 0.25 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr15_+_49170281 | 0.25 |
ENST00000560490.1 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chrX_+_12993202 | 0.24 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr7_-_100888313 | 0.23 |
ENST00000442303.1 |
FIS1 |
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr11_+_119039414 | 0.22 |
ENST00000409991.1 ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1 |
NLR family member X1 |
| chr17_+_68071458 | 0.21 |
ENST00000589377.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_54813229 | 0.20 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr19_+_42212526 | 0.20 |
ENST00000598976.1 ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA CEACAM5 |
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr9_-_14314518 | 0.19 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr10_-_124639062 | 0.19 |
ENST00000368898.3 ENST00000368896.1 ENST00000545804.1 |
FAM24B CUZD1 |
family with sequence similarity 24, member B CUB and zona pellucida-like domains 1 |
| chr5_-_137090028 | 0.18 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr3_-_49131013 | 0.17 |
ENST00000424300.1 |
QRICH1 |
glutamine-rich 1 |
| chr19_+_45254529 | 0.16 |
ENST00000444487.1 |
BCL3 |
B-cell CLL/lymphoma 3 |
| chr3_-_9834375 | 0.16 |
ENST00000343450.2 ENST00000301964.2 |
TADA3 |
transcriptional adaptor 3 |
| chr17_-_15466742 | 0.13 |
ENST00000584811.1 ENST00000419890.2 |
TVP23C |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr4_+_71588372 | 0.13 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr12_+_110011571 | 0.13 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr20_-_23807358 | 0.12 |
ENST00000304725.2 |
CST2 |
cystatin SA |
| chr20_-_23731569 | 0.12 |
ENST00000304749.2 |
CST1 |
cystatin SN |
| chr17_-_40950698 | 0.11 |
ENST00000328434.7 |
COA3 |
cytochrome c oxidase assembly factor 3 |
| chr18_-_52989217 | 0.11 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr17_+_68071389 | 0.11 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_-_10955226 | 0.11 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr18_-_52989525 | 0.10 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr13_+_45694583 | 0.09 |
ENST00000340473.6 |
GTF2F2 |
general transcription factor IIF, polypeptide 2, 30kDa |
| chr12_+_69186125 | 0.09 |
ENST00000399333.3 |
AC124890.1 |
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr19_+_49999631 | 0.07 |
ENST00000270625.2 ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11 |
ribosomal protein S11 |
| chrX_+_135579670 | 0.07 |
ENST00000218364.4 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
| chr6_-_31926208 | 0.07 |
ENST00000454913.1 ENST00000436289.2 |
NELFE |
negative elongation factor complex member E |
| chr17_+_38083977 | 0.06 |
ENST00000578802.1 ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4 |
RP11-387H17.4 |
| chr17_-_15466850 | 0.06 |
ENST00000438826.3 ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C TVP23C-CDRT4 |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr8_-_99954788 | 0.06 |
ENST00000523601.1 |
STK3 |
serine/threonine kinase 3 |
| chr12_+_101988627 | 0.06 |
ENST00000547405.1 ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr3_-_48594248 | 0.05 |
ENST00000545984.1 ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr1_-_6259641 | 0.05 |
ENST00000234875.4 |
RPL22 |
ribosomal protein L22 |
| chrX_-_41782249 | 0.04 |
ENST00000442742.2 ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr7_+_1127723 | 0.03 |
ENST00000397088.3 |
GPER1 |
G protein-coupled estrogen receptor 1 |
| chr4_-_83483395 | 0.03 |
ENST00000515780.2 |
TMEM150C |
transmembrane protein 150C |
| chr11_-_8190534 | 0.03 |
ENST00000309737.6 ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
| chr7_+_120969045 | 0.03 |
ENST00000222462.2 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
| chr2_+_200820494 | 0.03 |
ENST00000435773.2 |
C2orf47 |
chromosome 2 open reading frame 47 |
| chr9_-_34589700 | 0.02 |
ENST00000351266.4 |
CNTFR |
ciliary neurotrophic factor receptor |
| chr5_-_35230434 | 0.02 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr22_-_44208070 | 0.02 |
ENST00000396231.2 |
EFCAB6 |
EF-hand calcium binding domain 6 |
| chr3_+_185000729 | 0.02 |
ENST00000448876.1 ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr16_+_2079637 | 0.02 |
ENST00000561844.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr16_+_10971037 | 0.01 |
ENST00000324288.8 ENST00000381835.5 |
CIITA |
class II, major histocompatibility complex, transactivator |
| chr7_+_95401851 | 0.01 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr4_+_74718906 | 0.01 |
ENST00000226524.3 |
PF4V1 |
platelet factor 4 variant 1 |
| chr16_+_2079501 | 0.00 |
ENST00000563587.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr16_-_67514982 | 0.00 |
ENST00000565835.1 ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr10_-_135122603 | 0.00 |
ENST00000368563.2 |
TUBGCP2 |
tubulin, gamma complex associated protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.7 | 2.7 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.5 | 1.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.4 | 1.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.3 | 1.0 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 1.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 2.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 0.8 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 2.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 1.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 4.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.6 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.7 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 2.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.5 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 3.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 1.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 2.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:2000790 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 3.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 3.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 2.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.6 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.2 | 1.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 2.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 1.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |