Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NFATC1
Z-value: 1.41
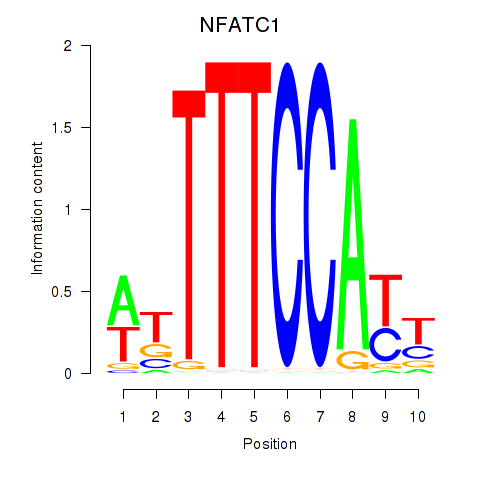
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | NFATC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg19_v2_chr18_+_77155856_77155939 | 0.19 | 4.8e-01 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_35691017 | 4.17 |
ENST00000378292.3 |
TPM2 |
tropomyosin 2 (beta) |
| chr3_-_127541194 | 4.16 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr1_-_153517473 | 4.15 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr3_-_158390282 | 3.07 |
ENST00000264265.3 |
LXN |
latexin |
| chr11_-_102668879 | 2.99 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr18_-_53177984 | 2.88 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr18_-_53257027 | 2.82 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr12_-_50616122 | 2.80 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr17_-_39780819 | 2.80 |
ENST00000311208.8 |
KRT17 |
keratin 17 |
| chr12_-_50616382 | 2.72 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr22_+_38071615 | 2.53 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr1_+_86046433 | 2.50 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr12_-_52887034 | 2.40 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr3_+_69134080 | 2.33 |
ENST00000273258.3 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_-_226926864 | 2.32 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr13_+_73629107 | 2.29 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr3_+_69134124 | 2.23 |
ENST00000478935.1 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr18_-_53089723 | 2.08 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr2_+_189839046 | 2.07 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr1_+_79115503 | 1.93 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr4_-_72649763 | 1.90 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr1_-_186649543 | 1.90 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_-_13835147 | 1.89 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr1_+_46640750 | 1.89 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr11_-_122930121 | 1.88 |
ENST00000524552.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr12_+_96588143 | 1.87 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr3_-_107777208 | 1.80 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr17_-_39743139 | 1.78 |
ENST00000167586.6 |
KRT14 |
keratin 14 |
| chr9_-_14314518 | 1.74 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr3_-_111314230 | 1.71 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr5_+_140729649 | 1.71 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr10_+_11206925 | 1.69 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr9_-_14314566 | 1.68 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr17_-_39677971 | 1.66 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr10_-_121296045 | 1.61 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr1_-_94703118 | 1.49 |
ENST00000260526.6 ENST00000370217.3 |
ARHGAP29 |
Rho GTPase activating protein 29 |
| chr5_-_94417339 | 1.46 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr4_+_146403912 | 1.45 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chrX_-_10544942 | 1.41 |
ENST00000380779.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr11_-_122929699 | 1.40 |
ENST00000526686.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr17_+_32582293 | 1.38 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr18_-_52989217 | 1.38 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr10_-_105212059 | 1.37 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr18_-_52989525 | 1.35 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_-_161349909 | 1.30 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr6_-_32920794 | 1.28 |
ENST00000395305.3 ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA XXbac-BPG181M17.5 |
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chrX_+_102611373 | 1.24 |
ENST00000372661.3 ENST00000372656.3 |
WBP5 |
WW domain binding protein 5 |
| chr8_+_95653302 | 1.24 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr5_+_49963239 | 1.24 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_95653427 | 1.24 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr10_-_105212141 | 1.23 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr4_-_71532339 | 1.22 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_+_95653373 | 1.21 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr10_+_13141441 | 1.20 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr7_+_134551583 | 1.20 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr5_+_140762268 | 1.19 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr1_-_103574024 | 1.19 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr1_-_111743285 | 1.17 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chr11_+_131240373 | 1.17 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr17_-_29648761 | 1.15 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr6_+_32407619 | 1.14 |
ENST00000395388.2 ENST00000374982.5 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
| chr12_-_91573249 | 1.12 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr14_-_30396802 | 1.10 |
ENST00000415220.2 |
PRKD1 |
protein kinase D1 |
| chr16_+_15528332 | 1.07 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr10_+_103825080 | 1.05 |
ENST00000299238.5 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
| chr5_+_49962495 | 1.05 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr12_-_91573316 | 1.05 |
ENST00000393155.1 |
DCN |
decorin |
| chr2_-_211168332 | 1.04 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr11_-_102651343 | 1.03 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr9_+_124062071 | 1.03 |
ENST00000373818.4 |
GSN |
gelsolin |
| chr2_+_33701286 | 1.02 |
ENST00000403687.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_+_70404996 | 1.02 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr11_-_14358620 | 1.00 |
ENST00000531421.1 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
| chr10_+_13141585 | 0.99 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr9_-_16870704 | 0.98 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr5_+_49962772 | 0.98 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr5_-_88179302 | 0.96 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_-_211179883 | 0.95 |
ENST00000352451.3 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr10_+_13142075 | 0.94 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr3_+_88188254 | 0.93 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr1_+_218519577 | 0.92 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr12_+_54447637 | 0.90 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr6_-_134495992 | 0.89 |
ENST00000475719.2 ENST00000367857.5 ENST00000237305.7 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chr15_+_33010175 | 0.88 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr22_-_19512893 | 0.88 |
ENST00000403084.1 ENST00000413119.2 |
CLDN5 |
claudin 5 |
| chr19_+_10197463 | 0.87 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr5_+_140810132 | 0.87 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr15_+_52155001 | 0.80 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr12_-_91573132 | 0.80 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr3_+_133293278 | 0.78 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr5_+_148521136 | 0.78 |
ENST00000506113.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr5_+_148521046 | 0.78 |
ENST00000326685.7 ENST00000356541.3 ENST00000309868.7 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chrX_-_13835461 | 0.77 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr14_+_90864504 | 0.77 |
ENST00000544280.2 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr4_-_159094194 | 0.76 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr6_+_129204337 | 0.74 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr8_-_122653630 | 0.74 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr20_-_14318248 | 0.72 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr9_+_27109133 | 0.72 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr14_-_92413353 | 0.71 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr10_-_92681033 | 0.71 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr4_+_41614909 | 0.71 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr12_+_96588279 | 0.70 |
ENST00000552142.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr4_+_26585686 | 0.70 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr7_+_107220422 | 0.70 |
ENST00000005259.4 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr11_-_111794446 | 0.69 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr15_-_43785303 | 0.69 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr3_-_136471204 | 0.69 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr2_+_234668894 | 0.69 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_162602244 | 0.69 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr4_-_38806404 | 0.68 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr15_+_63354769 | 0.66 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr16_+_1583567 | 0.65 |
ENST00000566264.1 |
TMEM204 |
transmembrane protein 204 |
| chr20_+_36149602 | 0.65 |
ENST00000062104.2 ENST00000346199.2 |
NNAT |
neuronatin |
| chr4_-_11431389 | 0.65 |
ENST00000002596.5 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr4_-_104021009 | 0.64 |
ENST00000509245.1 ENST00000296424.4 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
| chr14_-_30396948 | 0.64 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr18_+_29027696 | 0.64 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr12_-_28125638 | 0.61 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr20_+_55204351 | 0.58 |
ENST00000201031.2 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr11_-_102401469 | 0.58 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr1_+_209941942 | 0.58 |
ENST00000487271.1 ENST00000477431.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr11_-_110167352 | 0.58 |
ENST00000533991.1 ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX |
radixin |
| chr13_-_67804445 | 0.56 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr1_-_150780757 | 0.56 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr5_-_73937244 | 0.55 |
ENST00000302351.4 ENST00000510316.1 ENST00000508331.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr6_-_136871957 | 0.54 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr10_-_27149851 | 0.53 |
ENST00000376142.2 ENST00000359188.4 ENST00000376139.2 ENST00000376160.1 |
ABI1 |
abl-interactor 1 |
| chr3_+_130569592 | 0.53 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr11_-_2182388 | 0.53 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr5_+_135394840 | 0.50 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr12_-_52685312 | 0.49 |
ENST00000327741.5 |
KRT81 |
keratin 81 |
| chr12_+_20968608 | 0.48 |
ENST00000381541.3 ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3 SLCO1B3 SLCO1B7 |
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_+_132316081 | 0.48 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr4_+_154387480 | 0.48 |
ENST00000409663.3 ENST00000440693.1 ENST00000409959.3 |
KIAA0922 |
KIAA0922 |
| chr6_-_128841503 | 0.48 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr5_-_111093759 | 0.47 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr10_-_27149792 | 0.47 |
ENST00000376140.3 ENST00000376170.4 |
ABI1 |
abl-interactor 1 |
| chr12_+_27396901 | 0.46 |
ENST00000541191.1 ENST00000389032.3 |
STK38L |
serine/threonine kinase 38 like |
| chr7_+_55177416 | 0.45 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chrX_+_152760397 | 0.45 |
ENST00000331595.4 ENST00000431891.1 |
BGN |
biglycan |
| chr9_-_20622478 | 0.44 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr12_-_123756781 | 0.44 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr10_+_104535994 | 0.42 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr11_-_27723158 | 0.41 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr10_-_27149904 | 0.41 |
ENST00000376166.1 ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1 |
abl-interactor 1 |
| chr3_-_27498235 | 0.40 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_+_71561630 | 0.40 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr8_-_95274536 | 0.40 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr15_-_59981479 | 0.40 |
ENST00000607373.1 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr12_-_123756687 | 0.39 |
ENST00000261692.2 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr1_-_43638168 | 0.38 |
ENST00000431635.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr10_+_105036909 | 0.38 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr12_+_32260085 | 0.37 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr15_+_23255242 | 0.37 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr12_-_16759711 | 0.37 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_130569429 | 0.37 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr13_-_108867101 | 0.36 |
ENST00000356922.4 |
LIG4 |
ligase IV, DNA, ATP-dependent |
| chr14_-_51562745 | 0.36 |
ENST00000298355.3 |
TRIM9 |
tripartite motif containing 9 |
| chr12_+_131356582 | 0.35 |
ENST00000448750.3 ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN |
RAN, member RAS oncogene family |
| chr5_+_32788945 | 0.35 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr1_+_244214577 | 0.34 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr14_-_107013465 | 0.33 |
ENST00000390625.2 |
IGHV3-49 |
immunoglobulin heavy variable 3-49 |
| chr1_+_86934526 | 0.33 |
ENST00000394711.1 |
CLCA1 |
chloride channel accessory 1 |
| chr3_-_119813264 | 0.32 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr1_+_203734296 | 0.32 |
ENST00000442561.2 ENST00000367217.5 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
| chr6_+_63921351 | 0.31 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr9_-_215744 | 0.30 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr7_-_22862406 | 0.30 |
ENST00000372879.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr12_-_7656357 | 0.29 |
ENST00000396620.3 ENST00000432237.2 ENST00000359156.4 |
CD163 |
CD163 molecule |
| chr10_+_128593978 | 0.29 |
ENST00000280333.6 |
DOCK1 |
dedicator of cytokinesis 1 |
| chr1_-_159684371 | 0.29 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr4_-_178363581 | 0.28 |
ENST00000264595.2 |
AGA |
aspartylglucosaminidase |
| chr1_-_155880672 | 0.28 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr12_+_104359641 | 0.28 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr3_+_152017360 | 0.27 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr22_-_41215291 | 0.26 |
ENST00000542412.1 ENST00000544408.1 |
SLC25A17 |
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr17_+_27047570 | 0.26 |
ENST00000472628.1 ENST00000578181.1 |
RPL23A |
ribosomal protein L23a |
| chrX_-_63005405 | 0.25 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_+_234580499 | 0.25 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr2_+_234580525 | 0.25 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_92632542 | 0.25 |
ENST00000409154.4 ENST00000370378.4 |
KIAA1107 |
KIAA1107 |
| chr7_-_44530479 | 0.24 |
ENST00000355451.7 |
NUDCD3 |
NudC domain containing 3 |
| chr3_+_46395219 | 0.24 |
ENST00000445132.2 ENST00000292301.4 |
CCR2 |
chemokine (C-C motif) receptor 2 |
| chr13_-_49975632 | 0.23 |
ENST00000457041.1 ENST00000355854.4 |
CAB39L |
calcium binding protein 39-like |
| chr18_+_61575200 | 0.23 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr7_-_83824169 | 0.23 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_-_36986534 | 0.23 |
ENST00000429976.2 ENST00000301807.6 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr12_+_26111823 | 0.22 |
ENST00000381352.3 ENST00000535907.1 ENST00000405154.2 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_+_948803 | 0.22 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr19_-_2427863 | 0.22 |
ENST00000215570.3 |
TIMM13 |
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr14_-_92413727 | 0.21 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr12_-_13529642 | 0.21 |
ENST00000318426.2 |
C12orf36 |
chromosome 12 open reading frame 36 |
| chr12_+_98987369 | 0.20 |
ENST00000401722.3 ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| chr22_-_41215328 | 0.20 |
ENST00000434185.1 ENST00000435456.2 |
SLC25A17 |
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr2_-_228244013 | 0.20 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chr9_+_27109392 | 0.20 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr20_-_50808236 | 0.20 |
ENST00000361387.2 |
ZFP64 |
ZFP64 zinc finger protein |
| chr12_+_104359614 | 0.20 |
ENST00000266775.9 ENST00000544861.1 |
TDG |
thymine-DNA glycosylase |
| chr12_+_6644443 | 0.20 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 4.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 6.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 10.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 1.9 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.4 | 2.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 4.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 3.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 1.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.7 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 4.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 1.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 3.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.5 | GO:0051185 | FAD transmembrane transporter activity(GO:0015230) coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 5.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 2.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 7.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 2.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 1.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 6.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 4.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.7 | 3.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.6 | 2.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.6 | 2.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.6 | 4.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 3.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 1.9 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of platelet-derived growth factor production(GO:0090362) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.5 | 1.4 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.5 | 4.6 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.4 | 1.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 3.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 2.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.4 | 3.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.3 | 1.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 0.9 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 2.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 2.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 3.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 1.0 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 2.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.7 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 1.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.2 | 1.0 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.2 | 0.9 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.2 | 1.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 0.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.2 | 2.8 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 0.5 | GO:0043132 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) |
| 0.1 | 0.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 3.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 6.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.4 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.5 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.6 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.7 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.7 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.0 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 1.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 6.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 1.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.2 | GO:2000439 | monocyte extravasation(GO:0035696) negative regulation of eosinophil activation(GO:1902567) regulation of monocyte extravasation(GO:2000437) positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.7 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.0 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.2 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.4 | GO:0045955 | regulation of SNARE complex assembly(GO:0035542) negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cranial ganglion development(GO:0061550) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 3.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 9.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.6 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.7 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 2.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 4.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.9 | GO:0050821 | protein stabilization(GO:0050821) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 7.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 6.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 10.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.3 | 3.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 3.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 3.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 6.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 6.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 3.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 8.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 5.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |