Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
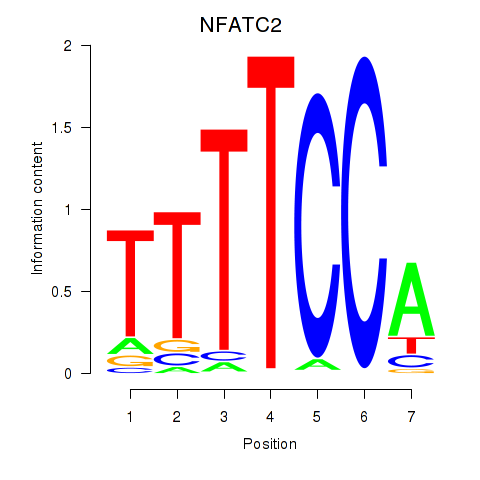
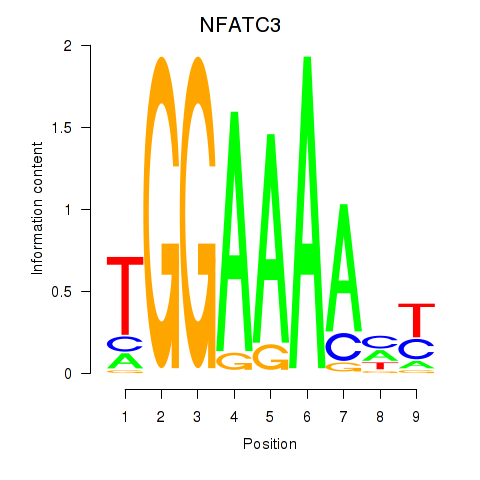
Results for NFATC2_NFATC3
Z-value: 2.16
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.15 | NFATC2 |
|
NFATC3
|
ENSG00000072736.14 | NFATC3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC3 | hg19_v2_chr16_+_68119247_68119293 | -0.64 | 7.7e-03 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190044480 | 8.41 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr9_-_35691017 | 7.72 |
ENST00000378292.3 |
TPM2 |
tropomyosin 2 (beta) |
| chr1_+_86046433 | 7.15 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr14_-_92413353 | 7.07 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr12_-_91573132 | 6.70 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573249 | 6.63 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr12_-_91573316 | 6.00 |
ENST00000393155.1 |
DCN |
decorin |
| chr1_-_153517473 | 5.73 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr8_-_13134045 | 5.69 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr2_-_163099885 | 5.44 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr2_-_163100045 | 5.23 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr2_-_238322770 | 5.20 |
ENST00000472056.1 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr11_+_101983176 | 5.15 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr2_-_238322800 | 4.75 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr5_+_135394840 | 4.73 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr14_-_92413727 | 4.63 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr2_-_238323007 | 4.30 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr8_+_70404996 | 4.27 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr11_-_119293872 | 3.88 |
ENST00000524970.1 |
THY1 |
Thy-1 cell surface antigen |
| chr14_-_52535712 | 3.88 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr12_-_91539918 | 3.68 |
ENST00000548218.1 |
DCN |
decorin |
| chr11_+_19799327 | 3.58 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr1_-_151965048 | 3.58 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr7_+_134551583 | 3.49 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr9_+_124062071 | 3.37 |
ENST00000373818.4 |
GSN |
gelsolin |
| chr1_-_68698222 | 3.28 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr6_+_86159821 | 3.27 |
ENST00000369651.3 |
NT5E |
5'-nucleotidase, ecto (CD73) |
| chr12_-_50616122 | 3.21 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr9_+_19049372 | 3.17 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr1_-_153538011 | 3.15 |
ENST00000368707.4 |
S100A2 |
S100 calcium binding protein A2 |
| chr1_-_150780757 | 3.13 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr8_-_18541603 | 3.11 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr8_+_98900132 | 3.05 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr6_+_86159765 | 3.03 |
ENST00000369646.3 ENST00000257770.3 |
NT5E |
5'-nucleotidase, ecto (CD73) |
| chr22_-_36236265 | 2.99 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_114035026 | 2.95 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr16_+_31483451 | 2.94 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr12_-_50616382 | 2.92 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr6_+_151662815 | 2.81 |
ENST00000359755.5 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr2_-_227664474 | 2.75 |
ENST00000305123.5 |
IRS1 |
insulin receptor substrate 1 |
| chr15_+_96869165 | 2.65 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr20_+_19867150 | 2.62 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr3_-_123512688 | 2.58 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr12_-_104443890 | 2.58 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr17_+_59477233 | 2.53 |
ENST00000240328.3 |
TBX2 |
T-box 2 |
| chrX_-_10544942 | 2.51 |
ENST00000380779.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr7_+_102553430 | 2.51 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr6_-_134495992 | 2.51 |
ENST00000475719.2 ENST00000367857.5 ENST00000237305.7 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chr2_-_161350305 | 2.45 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr15_+_63334831 | 2.41 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr7_-_41742697 | 2.38 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr1_-_94703118 | 2.37 |
ENST00000260526.6 ENST00000370217.3 |
ARHGAP29 |
Rho GTPase activating protein 29 |
| chr2_-_161349909 | 2.32 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr22_-_36220420 | 2.27 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_153518270 | 2.20 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr15_+_63340647 | 2.18 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr4_+_169418195 | 2.18 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr12_+_75874460 | 2.12 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_+_75874984 | 2.08 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_-_9268707 | 2.05 |
ENST00000318602.7 |
A2M |
alpha-2-macroglobulin |
| chr9_+_103204553 | 2.03 |
ENST00000502978.1 ENST00000334943.6 |
MSANTD3-TMEFF1 TMEFF1 |
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr12_+_26348429 | 2.01 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr15_-_90892669 | 1.98 |
ENST00000412799.2 |
GABARAPL3 |
GABA(A) receptors associated protein like 3, pseudogene |
| chr15_+_63354769 | 1.98 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr11_-_76381029 | 1.96 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr9_-_13165457 | 1.96 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr17_-_1389419 | 1.93 |
ENST00000575158.1 |
MYO1C |
myosin IC |
| chr20_-_34025999 | 1.92 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr8_-_23712312 | 1.91 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chr8_-_95274536 | 1.89 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr4_+_55095264 | 1.89 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_-_18480260 | 1.88 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr16_+_15528332 | 1.86 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr17_-_1389228 | 1.86 |
ENST00000438665.2 |
MYO1C |
myosin IC |
| chr15_+_63340858 | 1.84 |
ENST00000560615.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_63340734 | 1.81 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr4_-_114900831 | 1.80 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr12_+_75874580 | 1.78 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr1_+_82266053 | 1.77 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr10_-_62704005 | 1.72 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr15_+_63340775 | 1.69 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr13_-_33780133 | 1.68 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr4_-_143226979 | 1.64 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_76425368 | 1.64 |
ENST00000602540.1 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
| chr1_+_162602244 | 1.63 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr11_-_2950642 | 1.62 |
ENST00000314222.4 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
| chr5_+_32788945 | 1.62 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr1_-_68698197 | 1.61 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr2_+_46926326 | 1.61 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr9_-_13175823 | 1.55 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr5_+_155753745 | 1.54 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr5_-_111093759 | 1.52 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr16_+_56642041 | 1.49 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr4_+_41258786 | 1.49 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr7_+_79765071 | 1.49 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr11_-_111794446 | 1.48 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr6_-_52859046 | 1.48 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr11_+_19798964 | 1.48 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr5_-_42811986 | 1.45 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr4_+_169418255 | 1.43 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr22_-_36236623 | 1.43 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr7_+_55177416 | 1.42 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr9_+_137533615 | 1.40 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr1_-_156675368 | 1.36 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr5_+_140772381 | 1.35 |
ENST00000398604.2 |
PCDHGA8 |
protocadherin gamma subfamily A, 8 |
| chr10_-_105212059 | 1.35 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr1_-_149908710 | 1.34 |
ENST00000439741.2 ENST00000361405.6 ENST00000406732.3 |
MTMR11 |
myotubularin related protein 11 |
| chr13_+_102104980 | 1.33 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_+_99357319 | 1.33 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr12_+_96588143 | 1.32 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr15_+_32933866 | 1.30 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr5_-_42812143 | 1.28 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr10_+_128593978 | 1.26 |
ENST00000280333.6 |
DOCK1 |
dedicator of cytokinesis 1 |
| chr1_+_151512775 | 1.25 |
ENST00000368849.3 ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1 |
tuftelin 1 |
| chr6_-_112575912 | 1.25 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr5_+_119799927 | 1.23 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr1_-_156675535 | 1.22 |
ENST00000368221.1 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr12_+_26348246 | 1.21 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr6_+_17393839 | 1.21 |
ENST00000489374.1 ENST00000378990.2 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr19_-_36523709 | 1.20 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr2_+_201173667 | 1.20 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr18_-_500692 | 1.19 |
ENST00000400256.3 |
COLEC12 |
collectin sub-family member 12 |
| chr1_+_38022572 | 1.19 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr5_+_74011328 | 1.18 |
ENST00000513336.1 |
HEXB |
hexosaminidase B (beta polypeptide) |
| chrX_+_37208540 | 1.17 |
ENST00000466533.1 ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1 TM4SF2 |
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr2_-_190927447 | 1.16 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr7_+_120629653 | 1.16 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr11_+_131240373 | 1.15 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr1_+_244816237 | 1.15 |
ENST00000302550.11 |
DESI2 |
desumoylating isopeptidase 2 |
| chr10_-_127505167 | 1.14 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr1_+_26605618 | 1.14 |
ENST00000270792.5 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr6_+_17393888 | 1.13 |
ENST00000493172.1 ENST00000465994.1 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_-_111091948 | 1.13 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr12_-_49582978 | 1.12 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr11_-_117186946 | 1.12 |
ENST00000313005.6 ENST00000528053.1 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr18_+_3451584 | 1.10 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr10_-_105212141 | 1.08 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr18_+_66465302 | 1.07 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr6_-_167275991 | 1.07 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr7_-_151574191 | 1.07 |
ENST00000287878.4 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr6_+_158733692 | 1.07 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr14_+_24837226 | 1.07 |
ENST00000554050.1 ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr11_+_35211429 | 1.07 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr5_+_140186647 | 1.06 |
ENST00000512229.2 ENST00000356878.4 ENST00000530339.1 |
PCDHA4 |
protocadherin alpha 4 |
| chr20_+_43211149 | 1.06 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr12_+_10365404 | 1.06 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr3_-_73673991 | 1.05 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr14_-_30396948 | 1.05 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr5_-_77844974 | 1.02 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr17_-_46623441 | 1.01 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr19_+_2389784 | 1.00 |
ENST00000332578.3 |
TMPRSS9 |
transmembrane protease, serine 9 |
| chrX_+_37208521 | 0.99 |
ENST00000378628.4 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr8_-_11710979 | 0.98 |
ENST00000415599.2 |
CTSB |
cathepsin B |
| chr5_-_16742330 | 0.98 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr8_-_120685608 | 0.97 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chrX_-_17878827 | 0.96 |
ENST00000360011.1 |
RAI2 |
retinoic acid induced 2 |
| chr1_+_84609944 | 0.95 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_201170596 | 0.94 |
ENST00000439084.1 ENST00000409718.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr3_+_187930719 | 0.93 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_75273246 | 0.92 |
ENST00000526397.1 ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr14_+_21538517 | 0.92 |
ENST00000298693.3 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr5_+_140864649 | 0.92 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr5_+_140220769 | 0.91 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr9_+_35806082 | 0.91 |
ENST00000447210.1 |
NPR2 |
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr11_+_75273101 | 0.90 |
ENST00000533603.1 ENST00000358171.3 ENST00000526242.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr19_+_11658655 | 0.90 |
ENST00000588935.1 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr3_+_8543393 | 0.90 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr19_+_16296191 | 0.89 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr5_+_96211643 | 0.87 |
ENST00000437043.3 ENST00000510373.1 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
| chr1_-_32169761 | 0.84 |
ENST00000271069.6 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr1_-_32169920 | 0.82 |
ENST00000373672.3 ENST00000373668.3 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr6_+_39760783 | 0.82 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr9_+_115913222 | 0.80 |
ENST00000259392.3 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
| chr3_-_107941230 | 0.80 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chrX_-_102943022 | 0.80 |
ENST00000433176.2 |
MORF4L2 |
mortality factor 4 like 2 |
| chr2_+_201170770 | 0.80 |
ENST00000409988.3 ENST00000409385.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr12_+_79258444 | 0.80 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr5_+_140762268 | 0.79 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr8_+_94929969 | 0.77 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chrX_-_37706815 | 0.76 |
ENST00000378578.4 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
| chr1_+_46640750 | 0.76 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr4_-_41216492 | 0.76 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_+_99899180 | 0.76 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr3_+_38017264 | 0.76 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chrX_-_102942961 | 0.75 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr1_+_38022513 | 0.73 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr2_+_46926048 | 0.73 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr15_+_67358163 | 0.73 |
ENST00000327367.4 |
SMAD3 |
SMAD family member 3 |
| chr10_-_49860525 | 0.72 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr6_-_112575838 | 0.71 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr21_+_37442239 | 0.71 |
ENST00000530908.1 ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1 |
carbonyl reductase 1 |
| chr3_+_142442841 | 0.70 |
ENST00000476941.1 ENST00000273482.6 |
TRPC1 |
transient receptor potential cation channel, subfamily C, member 1 |
| chr19_+_51226573 | 0.70 |
ENST00000250340.4 |
CLEC11A |
C-type lectin domain family 11, member A |
| chr1_-_103574024 | 0.70 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr20_+_33759854 | 0.70 |
ENST00000216968.4 |
PROCR |
protein C receptor, endothelial |
| chr8_-_17555164 | 0.69 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr18_+_3447572 | 0.69 |
ENST00000548489.2 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr6_+_39760129 | 0.69 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr6_-_167276033 | 0.67 |
ENST00000503859.1 ENST00000506565.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr12_-_123756781 | 0.67 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 24.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 6.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 28.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 25.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 4.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 4.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 1.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.7 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 11.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 5.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 4.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.8 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 2.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.6 | 3.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.6 | 1.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 6.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.5 | 2.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 10.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 35.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 1.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 2.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 4.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 7.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.3 | 1.6 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.6 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 5.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 3.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 1.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 3.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 19.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 0.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 4.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 3.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.2 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.8 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 1.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 8.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 6.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 8.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 11.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 13.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 1.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.3 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 2.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 11.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 4.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 1.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0070679 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 2.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 2.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 1.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 1.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 4.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) peptide disulfide oxidoreductase activity(GO:0015037) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 4.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.0 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 5.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 2.5 | 9.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.9 | 23.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.6 | 4.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.6 | 6.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 1.5 | 11.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.3 | 3.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.3 | 5.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.1 | 6.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.0 | 9.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 1.0 | 11.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.8 | 2.5 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.8 | 2.4 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.7 | 3.4 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.7 | 2.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 5.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.6 | 1.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.6 | 1.9 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 1.9 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.6 | 3.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.6 | 4.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.5 | 1.6 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.5 | 2.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.5 | 1.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.5 | 0.5 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.5 | 1.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.4 | 1.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.4 | 5.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 2.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 5.0 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.4 | 1.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 | 2.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 1.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 1.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.3 | 1.3 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 2.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 0.6 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.3 | 1.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.3 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.3 | 0.8 | GO:0061317 | arterial endothelial cell fate commitment(GO:0060844) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.3 | 3.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 2.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 3.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 1.8 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.2 | 0.9 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 15.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 2.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 3.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.2 | 1.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 2.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 0.9 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.2 | 1.8 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 1.0 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.5 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.2 | 3.3 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.6 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.2 | 1.0 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 1.9 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.2 | 0.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 3.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 3.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.5 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.1 | GO:1900625 | monocyte aggregation(GO:0070487) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 3.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.4 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.5 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 1.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 1.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.1 | 1.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 6.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.7 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.4 | GO:0071306 | cellular response to vitamin E(GO:0071306) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.4 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 1.9 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 0.3 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 1.1 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 7.4 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.4 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.1 | 0.4 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 1.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.9 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.6 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.1 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.4 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 1.2 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 0.2 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 1.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 | 0.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.2 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.8 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.3 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 5.3 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 1.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 1.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 1.6 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 1.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:0044546 | NLRP3 inflammasome complex assembly(GO:0044546) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 2.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 1.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 1.2 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 1.3 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.2 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.3 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 1.4 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.3 | GO:0060393 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) pathway-restricted SMAD protein phosphorylation(GO:0060389) regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 28.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 20.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 16.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 6.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 15.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 5.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 5.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 10.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 3.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 7.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.7 | 37.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.2 | 11.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.0 | 19.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.0 | 5.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.0 | 10.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.9 | 3.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.8 | 2.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.6 | 4.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.6 | 2.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 3.2 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 1.4 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.5 | 6.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 4.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.3 | 4.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 1.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.2 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.7 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 3.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 5.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 3.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 14.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 8.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 8.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 3.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 5.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.2 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 6.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 5.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 9.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0097449 | eukaryotic translation initiation factor 2 complex(GO:0005850) astrocyte projection(GO:0097449) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |