Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
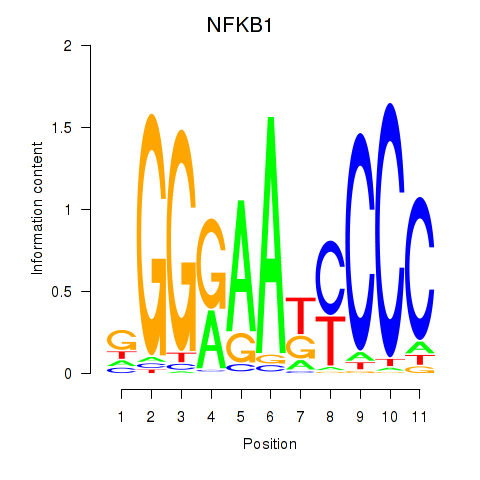
Results for NFKB1
Z-value: 1.89
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | NFKB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg19_v2_chr4_+_103423055_103423112 | 0.68 | 3.9e-03 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_149792295 | 9.70 |
ENST00000518797.1 ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr19_-_6591113 | 6.93 |
ENST00000423145.3 ENST00000245903.3 |
CD70 |
CD70 molecule |
| chr14_+_75988851 | 6.56 |
ENST00000555504.1 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr6_+_14117872 | 6.08 |
ENST00000379153.3 |
CD83 |
CD83 molecule |
| chr6_+_32605134 | 5.70 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_+_138188551 | 5.70 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr6_+_32605195 | 5.70 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr11_+_102188272 | 5.65 |
ENST00000532808.1 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr6_-_29527702 | 5.64 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr11_+_102188224 | 5.62 |
ENST00000263464.3 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr1_+_111770278 | 4.89 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr19_+_42381337 | 4.85 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr19_+_42381173 | 4.63 |
ENST00000221972.3 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr12_-_9913489 | 4.54 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr19_+_1067492 | 3.94 |
ENST00000586866.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr19_+_1067144 | 3.90 |
ENST00000313093.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr14_-_35873856 | 3.90 |
ENST00000553342.1 ENST00000216797.5 ENST00000557140.1 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr19_+_1067271 | 3.88 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr20_+_57430162 | 3.71 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr19_+_50922187 | 3.50 |
ENST00000595883.1 ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr22_-_37640456 | 3.40 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr10_+_30722866 | 3.38 |
ENST00000263056.1 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
| chr6_+_31540056 | 3.37 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr10_+_12391481 | 3.33 |
ENST00000378847.3 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
| chr7_+_69064300 | 3.27 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr1_+_111770232 | 3.26 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr14_+_75988768 | 3.23 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr6_-_31550192 | 3.12 |
ENST00000429299.2 ENST00000446745.2 |
LTB |
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_+_29691198 | 3.08 |
ENST00000440587.2 ENST00000434407.2 |
HLA-F |
major histocompatibility complex, class I, F |
| chr20_+_44746885 | 3.04 |
ENST00000372285.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr6_+_29691056 | 3.03 |
ENST00000414333.1 ENST00000334668.4 ENST00000259951.7 |
HLA-F |
major histocompatibility complex, class I, F |
| chr4_-_40631859 | 3.03 |
ENST00000295971.7 ENST00000319592.4 |
RBM47 |
RNA binding motif protein 47 |
| chr22_-_37640277 | 2.94 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr9_-_123691047 | 2.88 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr6_+_135502466 | 2.85 |
ENST00000367814.4 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr16_+_50776021 | 2.76 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr15_+_85923797 | 2.74 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr14_+_61789382 | 2.68 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr6_+_32821924 | 2.66 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_+_103422471 | 2.62 |
ENST00000226574.4 ENST00000394820.4 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr11_+_128562372 | 2.60 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_+_85923856 | 2.59 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr2_+_61108650 | 2.58 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr20_+_9494987 | 2.57 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr2_-_163175133 | 2.40 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr3_+_53195136 | 2.36 |
ENST00000394729.2 ENST00000330452.3 |
PRKCD |
protein kinase C, delta |
| chr16_+_50730910 | 2.32 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr20_+_44746939 | 2.30 |
ENST00000372276.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr1_-_202130702 | 2.30 |
ENST00000309017.3 ENST00000477554.1 ENST00000492451.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr6_-_44233361 | 2.25 |
ENST00000275015.5 |
NFKBIE |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr2_-_191885686 | 2.22 |
ENST00000432058.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr2_+_228678550 | 2.20 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr16_+_50775971 | 2.17 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr2_+_61108771 | 2.10 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr1_+_156123359 | 2.09 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr4_-_76944621 | 2.07 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr6_-_32821599 | 2.05 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_150466692 | 2.04 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr6_+_106534192 | 2.02 |
ENST00000369091.2 ENST00000369096.4 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr1_+_156123318 | 1.99 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_-_58345569 | 1.95 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr6_+_29910301 | 1.92 |
ENST00000376809.5 ENST00000376802.2 |
HLA-A |
major histocompatibility complex, class I, A |
| chr5_-_150460539 | 1.71 |
ENST00000520931.1 ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr12_+_11802753 | 1.70 |
ENST00000396373.4 |
ETV6 |
ets variant 6 |
| chr14_+_66975213 | 1.68 |
ENST00000543237.1 ENST00000305960.9 |
GPHN |
gephyrin |
| chr2_+_163175394 | 1.63 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr16_+_50775948 | 1.60 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chrX_-_73072534 | 1.57 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr19_+_2476116 | 1.57 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr10_+_104154229 | 1.55 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr11_-_46142948 | 1.54 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr17_-_7590745 | 1.53 |
ENST00000514944.1 ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53 |
tumor protein p53 |
| chr2_+_208394658 | 1.53 |
ENST00000421139.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr19_-_46234119 | 1.47 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chrX_-_30595959 | 1.45 |
ENST00000378962.3 |
CXorf21 |
chromosome X open reading frame 21 |
| chr19_+_496454 | 1.44 |
ENST00000346144.4 ENST00000215637.3 ENST00000382683.4 |
MADCAM1 |
mucosal vascular addressin cell adhesion molecule 1 |
| chr4_-_76928641 | 1.44 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr11_-_72853091 | 1.43 |
ENST00000311172.7 ENST00000409314.1 |
FCHSD2 |
FCH and double SH3 domains 2 |
| chr2_+_97203082 | 1.41 |
ENST00000454558.2 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr2_-_152955213 | 1.41 |
ENST00000427385.1 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_+_292051 | 1.38 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr9_-_115480303 | 1.33 |
ENST00000374234.1 ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP |
INTS3 and NABP interacting protein |
| chr5_-_150460914 | 1.33 |
ENST00000389378.2 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr4_-_185395672 | 1.33 |
ENST00000393593.3 |
IRF2 |
interferon regulatory factor 2 |
| chr2_-_152955537 | 1.32 |
ENST00000201943.5 ENST00000539935.1 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_+_208394616 | 1.30 |
ENST00000432329.2 ENST00000353267.3 ENST00000445803.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr5_-_150521192 | 1.27 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr3_-_4793274 | 1.23 |
ENST00000414938.1 |
EGOT |
eosinophil granule ontogeny transcript (non-protein coding) |
| chr19_-_2050852 | 1.22 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr2_+_38893047 | 1.21 |
ENST00000272252.5 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
| chr19_+_45504688 | 1.21 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_+_97202480 | 1.18 |
ENST00000357485.3 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr11_-_77185094 | 1.17 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr4_+_142558078 | 1.11 |
ENST00000529613.1 |
IL15 |
interleukin 15 |
| chr14_-_64970494 | 1.09 |
ENST00000608382.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr15_-_66084428 | 1.07 |
ENST00000443035.3 ENST00000431932.2 |
DENND4A |
DENN/MADD domain containing 4A |
| chr1_+_211432593 | 1.05 |
ENST00000367006.4 |
RCOR3 |
REST corepressor 3 |
| chr14_+_64970662 | 1.04 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr4_+_142557771 | 1.04 |
ENST00000514653.1 |
IL15 |
interleukin 15 |
| chr4_+_142557717 | 1.03 |
ENST00000320650.4 ENST00000296545.7 |
IL15 |
interleukin 15 |
| chr12_-_12849073 | 1.03 |
ENST00000332427.2 ENST00000540796.1 |
GPR19 |
G protein-coupled receptor 19 |
| chr14_+_103243813 | 1.03 |
ENST00000560371.1 ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3 |
TNF receptor-associated factor 3 |
| chr15_-_71055878 | 1.00 |
ENST00000322954.6 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr11_-_46142615 | 1.00 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr19_-_2042065 | 0.99 |
ENST00000591588.1 ENST00000591142.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr1_-_202129105 | 0.95 |
ENST00000367279.4 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chrX_-_112084043 | 0.95 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr2_+_208394455 | 0.93 |
ENST00000430624.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr12_+_52445191 | 0.90 |
ENST00000243050.1 ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
| chr14_-_24616426 | 0.88 |
ENST00000216802.5 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr1_-_202129704 | 0.86 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr10_-_97050777 | 0.85 |
ENST00000329399.6 |
PDLIM1 |
PDZ and LIM domain 1 |
| chr10_+_12391685 | 0.85 |
ENST00000378845.1 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
| chr6_+_144471643 | 0.84 |
ENST00000367568.4 |
STX11 |
syntaxin 11 |
| chr1_+_37940153 | 0.84 |
ENST00000373087.6 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
| chr5_-_96143602 | 0.79 |
ENST00000443439.2 ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr14_-_24615805 | 0.74 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr15_+_89346699 | 0.74 |
ENST00000558207.1 |
ACAN |
aggrecan |
| chr8_-_9008206 | 0.73 |
ENST00000310455.3 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
| chr4_-_140098339 | 0.73 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr1_-_54304212 | 0.72 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr11_+_63997750 | 0.71 |
ENST00000321685.3 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr19_-_39330818 | 0.69 |
ENST00000594769.1 ENST00000602021.1 |
AC104534.3 |
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chrX_-_15872914 | 0.68 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr3_+_57261743 | 0.66 |
ENST00000288266.3 |
APPL1 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr12_-_49259643 | 0.66 |
ENST00000309739.5 |
RND1 |
Rho family GTPase 1 |
| chr1_-_54303949 | 0.66 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr19_-_13617037 | 0.66 |
ENST00000360228.5 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr5_+_139781445 | 0.65 |
ENST00000532219.1 ENST00000394722.3 |
ANKHD1-EIF4EBP3 ANKHD1 |
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr5_+_139781393 | 0.65 |
ENST00000360839.2 ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
| chr4_-_103748696 | 0.64 |
ENST00000321805.7 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_7590734 | 0.63 |
ENST00000457584.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr1_-_41950342 | 0.62 |
ENST00000372587.4 |
EDN2 |
endothelin 2 |
| chr2_+_208394794 | 0.60 |
ENST00000536726.1 ENST00000374397.4 ENST00000452474.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr6_-_111804905 | 0.58 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr1_+_62417957 | 0.58 |
ENST00000307297.7 ENST00000543708.1 |
INADL |
InaD-like (Drosophila) |
| chr14_+_76618242 | 0.58 |
ENST00000557542.1 ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L |
G patch domain containing 2-like |
| chr1_-_161168834 | 0.58 |
ENST00000367995.3 ENST00000367996.5 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
| chr1_-_155658085 | 0.57 |
ENST00000311573.5 ENST00000438245.2 |
YY1AP1 |
YY1 associated protein 1 |
| chr1_-_173886491 | 0.57 |
ENST00000367698.3 |
SERPINC1 |
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr5_-_135231516 | 0.54 |
ENST00000274520.1 |
IL9 |
interleukin 9 |
| chr2_-_136743169 | 0.54 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr1_-_54303934 | 0.53 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr12_+_49761224 | 0.53 |
ENST00000553127.1 ENST00000321898.6 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr17_+_40440481 | 0.53 |
ENST00000590726.2 ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A |
signal transducer and activator of transcription 5A |
| chr2_-_100721178 | 0.52 |
ENST00000409236.2 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr11_-_3862206 | 0.51 |
ENST00000351018.4 |
RHOG |
ras homolog family member G |
| chr19_-_47734448 | 0.50 |
ENST00000439096.2 |
BBC3 |
BCL2 binding component 3 |
| chr21_+_34775698 | 0.49 |
ENST00000381995.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr2_-_100721923 | 0.49 |
ENST00000356421.2 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr12_+_57624119 | 0.48 |
ENST00000555773.1 ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_100881109 | 0.47 |
ENST00000308344.5 |
CLDN15 |
claudin 15 |
| chr8_+_104831554 | 0.45 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr3_+_9439400 | 0.43 |
ENST00000450326.1 ENST00000402198.1 ENST00000402466.1 |
SETD5 |
SET domain containing 5 |
| chr4_-_103748271 | 0.43 |
ENST00000343106.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_24439148 | 0.42 |
ENST00000543805.1 ENST00000534993.1 |
DHRS4L2 |
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr8_+_73449625 | 0.41 |
ENST00000523207.1 |
KCNB2 |
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr12_+_57624085 | 0.41 |
ENST00000553474.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_-_43205811 | 0.40 |
ENST00000372539.3 ENST00000296387.1 ENST00000539749.1 |
CLDN19 |
claudin 19 |
| chr16_-_10276611 | 0.40 |
ENST00000396573.2 |
GRIN2A |
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| chr12_+_57623869 | 0.39 |
ENST00000414700.3 ENST00000557703.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr20_-_62103862 | 0.39 |
ENST00000344462.4 ENST00000357249.2 ENST00000359125.2 ENST00000360480.3 ENST00000370224.1 ENST00000344425.5 ENST00000354587.3 ENST00000359689.1 |
KCNQ2 |
potassium voltage-gated channel, KQT-like subfamily, member 2 |
| chrX_+_147582228 | 0.39 |
ENST00000342251.3 |
AFF2 |
AF4/FMR2 family, member 2 |
| chr6_+_149068464 | 0.39 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr2_+_64681641 | 0.38 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr17_+_8924837 | 0.38 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr12_-_118810688 | 0.38 |
ENST00000542532.1 ENST00000392533.3 |
TAOK3 |
TAO kinase 3 |
| chr21_+_34775772 | 0.37 |
ENST00000405436.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr10_+_21823079 | 0.37 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr11_-_133826852 | 0.37 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr21_+_34775181 | 0.36 |
ENST00000290219.6 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr6_+_31949801 | 0.36 |
ENST00000428956.2 ENST00000498271.1 |
C4A |
complement component 4A (Rodgers blood group) |
| chr11_+_118307179 | 0.36 |
ENST00000534358.1 ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
| chr8_-_70747205 | 0.36 |
ENST00000260126.4 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr12_+_57623477 | 0.36 |
ENST00000557487.1 ENST00000555634.1 ENST00000556689.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr4_-_122085469 | 0.35 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr17_-_7154984 | 0.35 |
ENST00000574322.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr12_-_24103954 | 0.34 |
ENST00000441133.2 ENST00000545921.1 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr1_+_54359854 | 0.34 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr19_-_13617247 | 0.34 |
ENST00000573710.2 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr9_+_82188077 | 0.34 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr17_+_40996590 | 0.34 |
ENST00000253799.3 ENST00000452774.2 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
| chr11_-_44331679 | 0.33 |
ENST00000329255.3 |
ALX4 |
ALX homeobox 4 |
| chr12_+_56732658 | 0.33 |
ENST00000228534.4 |
IL23A |
interleukin 23, alpha subunit p19 |
| chr1_-_205744574 | 0.33 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr19_+_42364460 | 0.32 |
ENST00000593863.1 |
RPS19 |
ribosomal protein S19 |
| chr3_+_9439579 | 0.32 |
ENST00000406341.1 |
SETD5 |
SET domain containing 5 |
| chr17_+_38465441 | 0.29 |
ENST00000577646.1 ENST00000254066.5 |
RARA |
retinoic acid receptor, alpha |
| chr19_-_49149553 | 0.29 |
ENST00000084798.4 |
CA11 |
carbonic anhydrase XI |
| chr12_-_6716569 | 0.29 |
ENST00000544040.1 ENST00000545942.1 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
| chr11_-_64570706 | 0.27 |
ENST00000294066.2 ENST00000377350.3 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr8_+_104831472 | 0.27 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr7_+_143013198 | 0.27 |
ENST00000343257.2 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
| chr4_+_114214125 | 0.26 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chr12_-_57824739 | 0.26 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chrX_-_119709637 | 0.26 |
ENST00000404115.3 |
CUL4B |
cullin 4B |
| chr1_-_57889687 | 0.26 |
ENST00000371236.2 ENST00000371230.1 |
DAB1 |
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr4_-_5891918 | 0.25 |
ENST00000512574.1 |
CRMP1 |
collapsin response mediator protein 1 |
| chr1_-_161279749 | 0.25 |
ENST00000533357.1 ENST00000360451.6 ENST00000336559.4 |
MPZ |
myelin protein zero |
| chr1_-_205744205 | 0.24 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.4 | 9.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.2 | 14.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.8 | 5.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 7.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 1.9 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.6 | 4.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.6 | 11.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.4 | 2.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 2.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 2.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 0.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 6.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 5.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 5.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 3.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 2.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 6.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 4.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 9.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 3.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 6.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 25.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.5 | 1.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.5 | 11.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 3.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 9.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 3.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 4.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 3.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 1.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 6.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 2.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 7.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 9.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 10.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 9.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 5.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.1 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 32.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 7.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 8.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 7.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 4.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 1.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 4.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 2.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 3.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 8.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.4 | 8.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.2 | 12.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.0 | 5.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.9 | 11.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.7 | 3.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.7 | 8.2 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.7 | 2.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 4.4 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.5 | 1.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 5.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 1.7 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.4 | 18.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 3.7 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 5.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 2.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 1.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 4.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 10.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 1.6 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 2.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 10.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 5.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 6.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 5.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 3.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.4 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 3.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 4.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 10.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 2.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 2.2 | 6.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.9 | 9.7 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.9 | 5.7 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.8 | 5.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.7 | 5.1 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.3 | 3.9 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 1.3 | 11.3 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 1.1 | 5.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.1 | 3.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 1.1 | 3.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 1.0 | 2.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 1.0 | 2.9 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.8 | 5.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.8 | 3.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.8 | 5.3 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.8 | 6.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.7 | 8.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 7.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.7 | 2.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.7 | 4.4 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.7 | 10.7 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.6 | 1.9 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.6 | 2.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.6 | 2.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.6 | 1.7 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.6 | 2.2 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.5 | 10.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.5 | 3.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 2.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.5 | 1.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.5 | 3.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 2.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 1.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 1.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 2.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.3 | 5.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.3 | 0.6 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 | 1.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 0.8 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 1.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.3 | 1.0 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 1.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 1.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 2.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.2 | 0.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 1.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.5 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.2 | 1.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.2 | 1.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 1.0 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 2.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 2.8 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 14.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.4 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 2.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 8.9 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.0 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 11.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 1.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.8 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.4 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) |
| 0.0 | 0.2 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.4 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 3.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 2.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 2.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.7 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 2.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 2.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 1.2 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) cranial skeletal system development(GO:1904888) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.0 | GO:0060167 | regulation of axon diameter(GO:0031133) regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |