Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
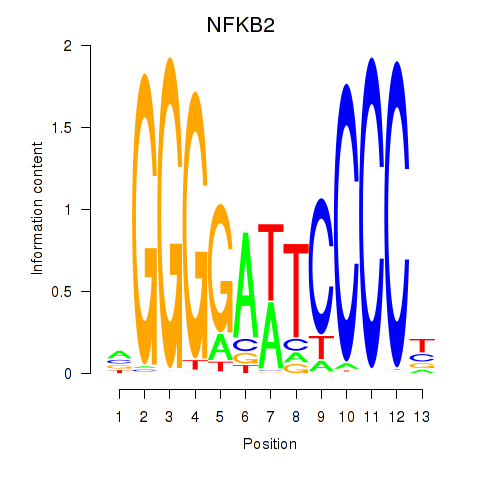
Results for NFKB2
Z-value: 0.91
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.13 | NFKB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg19_v2_chr10_+_104155450_104155479 | 0.84 | 4.6e-05 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_209824643 | 3.09 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr22_-_37640277 | 1.98 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr16_+_81812863 | 1.90 |
ENST00000359376.3 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr6_+_32605195 | 1.87 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_+_32605134 | 1.86 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr1_+_111770278 | 1.86 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr6_+_14117872 | 1.84 |
ENST00000379153.3 |
CD83 |
CD83 molecule |
| chr1_+_111770232 | 1.82 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr22_-_37640456 | 1.72 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr8_+_54793454 | 1.63 |
ENST00000276500.4 |
RGS20 |
regulator of G-protein signaling 20 |
| chr8_+_54793425 | 1.62 |
ENST00000522225.1 |
RGS20 |
regulator of G-protein signaling 20 |
| chr9_-_123691047 | 1.53 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr18_+_21529811 | 1.47 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr19_+_42381173 | 1.42 |
ENST00000221972.3 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr1_-_27961720 | 1.40 |
ENST00000545953.1 ENST00000374005.3 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr19_-_6591113 | 1.39 |
ENST00000423145.3 ENST00000245903.3 |
CD70 |
CD70 molecule |
| chr19_+_42381337 | 1.36 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr20_+_49411543 | 1.25 |
ENST00000609336.1 ENST00000445038.1 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr1_-_183559693 | 1.24 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr11_-_57335750 | 1.22 |
ENST00000340573.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr1_-_183560011 | 1.17 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr17_+_72462525 | 1.15 |
ENST00000360141.3 |
CD300A |
CD300a molecule |
| chr6_+_106959718 | 1.09 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chr17_-_34207295 | 1.07 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr12_+_7055631 | 1.04 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_53195136 | 1.02 |
ENST00000394729.2 ENST00000330452.3 |
PRKCD |
protein kinase C, delta |
| chr17_+_72462766 | 1.01 |
ENST00000392625.3 ENST00000361933.3 ENST00000310828.5 |
CD300A |
CD300a molecule |
| chr17_-_39928106 | 0.95 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr6_-_31324943 | 0.94 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr6_+_31540056 | 0.93 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr8_+_86376081 | 0.92 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr11_+_67351019 | 0.89 |
ENST00000398606.3 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr17_+_76164639 | 0.89 |
ENST00000225777.3 ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2 |
synaptogyrin 2 |
| chr16_+_50776021 | 0.85 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr17_+_76165213 | 0.82 |
ENST00000590201.1 |
SYNGR2 |
synaptogyrin 2 |
| chr20_+_49411431 | 0.76 |
ENST00000358791.5 ENST00000262591.5 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr20_+_43803517 | 0.74 |
ENST00000243924.3 |
PI3 |
peptidase inhibitor 3, skin-derived |
| chr2_+_61108650 | 0.74 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr11_+_67351213 | 0.72 |
ENST00000398603.1 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr2_-_89292422 | 0.69 |
ENST00000495489.1 |
IGKV1-8 |
immunoglobulin kappa variable 1-8 |
| chr3_-_56717246 | 0.68 |
ENST00000355628.5 |
FAM208A |
family with sequence similarity 208, member A |
| chr20_+_49411523 | 0.68 |
ENST00000371608.2 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr1_+_65613217 | 0.66 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr2_+_61108771 | 0.64 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr5_+_67511524 | 0.61 |
ENST00000521381.1 ENST00000521657.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr2_+_64681641 | 0.60 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr10_+_112257596 | 0.60 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr3_+_52232102 | 0.59 |
ENST00000469224.1 ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr16_+_15737124 | 0.58 |
ENST00000396355.1 ENST00000396353.2 |
NDE1 |
nudE neurodevelopment protein 1 |
| chr1_-_209979375 | 0.57 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr16_-_68269971 | 0.57 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr6_+_135502408 | 0.57 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_+_706113 | 0.55 |
ENST00000318562.8 ENST00000533256.1 ENST00000534755.1 |
EPS8L2 |
EPS8-like 2 |
| chr20_-_4795747 | 0.52 |
ENST00000379376.2 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr10_+_89622870 | 0.52 |
ENST00000371953.3 |
PTEN |
phosphatase and tensin homolog |
| chrX_+_49028265 | 0.50 |
ENST00000376322.3 ENST00000376327.5 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
| chr11_+_58910201 | 0.50 |
ENST00000528737.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr10_-_36813162 | 0.48 |
ENST00000440465.1 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
| chr1_+_42922173 | 0.48 |
ENST00000455780.1 ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr1_+_42921761 | 0.47 |
ENST00000372562.1 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr16_+_50775971 | 0.47 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr4_+_4387983 | 0.46 |
ENST00000397958.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chrX_+_64887512 | 0.45 |
ENST00000360270.5 |
MSN |
moesin |
| chr2_+_219081817 | 0.45 |
ENST00000315717.5 ENST00000420104.1 ENST00000295685.10 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr16_+_2867164 | 0.44 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr14_+_66975213 | 0.44 |
ENST00000543237.1 ENST00000305960.9 |
GPHN |
gephyrin |
| chr1_-_182573514 | 0.44 |
ENST00000367558.5 |
RGS16 |
regulator of G-protein signaling 16 |
| chr3_-_182698381 | 0.44 |
ENST00000292782.4 |
DCUN1D1 |
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr11_-_104840093 | 0.43 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr1_+_2487800 | 0.43 |
ENST00000355716.4 |
TNFRSF14 |
tumor necrosis factor receptor superfamily, member 14 |
| chr2_+_74056066 | 0.43 |
ENST00000339566.3 ENST00000409707.1 ENST00000452725.1 ENST00000432295.2 ENST00000424659.1 ENST00000394073.1 |
STAMBP |
STAM binding protein |
| chr2_+_74056147 | 0.43 |
ENST00000394070.2 ENST00000536064.1 |
STAMBP |
STAM binding protein |
| chrX_+_47050236 | 0.42 |
ENST00000377351.4 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr11_+_64949158 | 0.42 |
ENST00000527739.1 ENST00000526966.1 ENST00000533129.1 ENST00000524773.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr15_-_43559055 | 0.41 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr14_-_24615805 | 0.41 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr15_+_92396920 | 0.41 |
ENST00000318445.6 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr20_+_4667094 | 0.40 |
ENST00000424424.1 ENST00000457586.1 |
PRNP |
prion protein |
| chr11_+_66247478 | 0.40 |
ENST00000531863.1 ENST00000532677.1 |
DPP3 |
dipeptidyl-peptidase 3 |
| chr3_+_23847394 | 0.40 |
ENST00000306627.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr9_+_130911770 | 0.40 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr1_-_205744574 | 0.39 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr2_-_70781087 | 0.39 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr13_-_111214015 | 0.39 |
ENST00000267328.3 |
RAB20 |
RAB20, member RAS oncogene family |
| chr3_+_111718173 | 0.39 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr1_-_183604794 | 0.39 |
ENST00000367534.1 ENST00000359856.6 ENST00000294742.6 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
| chrX_+_150151824 | 0.38 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr3_+_23847432 | 0.38 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr11_+_58910295 | 0.37 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr5_-_150466692 | 0.37 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr11_+_64949343 | 0.37 |
ENST00000279247.6 ENST00000532285.1 ENST00000534373.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr21_+_27011584 | 0.37 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr9_+_130911723 | 0.37 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr2_-_38604398 | 0.37 |
ENST00000443098.1 ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2 |
atlastin GTPase 2 |
| chr1_-_40042416 | 0.37 |
ENST00000372857.3 ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4 |
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr14_+_97263641 | 0.36 |
ENST00000216639.3 |
VRK1 |
vaccinia related kinase 1 |
| chr11_+_66247880 | 0.36 |
ENST00000360510.2 ENST00000453114.1 ENST00000541961.1 ENST00000532019.1 ENST00000526515.1 ENST00000530165.1 ENST00000533725.1 |
DPP3 |
dipeptidyl-peptidase 3 |
| chr16_+_2867228 | 0.36 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr10_+_64133934 | 0.36 |
ENST00000395254.3 ENST00000395255.3 ENST00000410046.3 |
ZNF365 |
zinc finger protein 365 |
| chr4_-_169239921 | 0.36 |
ENST00000514995.1 ENST00000393743.3 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr1_-_93426998 | 0.35 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chrX_+_103173457 | 0.34 |
ENST00000419165.1 |
TMSB15B |
thymosin beta 15B |
| chr6_+_149068464 | 0.34 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr19_+_45504688 | 0.33 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_+_64949899 | 0.33 |
ENST00000531068.1 ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr15_+_85923856 | 0.32 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr8_+_22857048 | 0.32 |
ENST00000251822.6 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr16_+_50775948 | 0.32 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr17_+_66244071 | 0.32 |
ENST00000580548.1 ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr1_-_205744205 | 0.32 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr3_+_111718036 | 0.31 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr21_+_27011899 | 0.31 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr19_+_30302805 | 0.31 |
ENST00000262643.3 ENST00000575243.1 ENST00000357943.5 |
CCNE1 |
cyclin E1 |
| chr1_-_202129105 | 0.31 |
ENST00000367279.4 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr2_-_89340242 | 0.31 |
ENST00000480492.1 |
IGKV1-12 |
immunoglobulin kappa variable 1-12 |
| chr15_+_92397051 | 0.31 |
ENST00000424469.2 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr18_-_19283649 | 0.30 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr6_-_143266297 | 0.30 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr19_-_41859814 | 0.29 |
ENST00000221930.5 |
TGFB1 |
transforming growth factor, beta 1 |
| chr19_+_48824711 | 0.29 |
ENST00000599704.1 |
EMP3 |
epithelial membrane protein 3 |
| chr19_-_10450328 | 0.28 |
ENST00000160262.5 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr21_-_45079341 | 0.28 |
ENST00000443485.1 ENST00000291560.2 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr3_+_51705222 | 0.27 |
ENST00000457573.1 ENST00000341333.5 ENST00000412249.1 ENST00000425781.1 ENST00000415259.1 ENST00000395057.1 ENST00000416589.1 |
TEX264 |
testis expressed 264 |
| chrX_-_2418596 | 0.27 |
ENST00000381218.3 |
ZBED1 |
zinc finger, BED-type containing 1 |
| chr1_-_202129704 | 0.27 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr15_+_85923797 | 0.26 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr8_-_130951940 | 0.26 |
ENST00000522250.1 ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr9_+_100745615 | 0.26 |
ENST00000339399.4 |
ANP32B |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr19_+_10216899 | 0.25 |
ENST00000428358.1 ENST00000393796.4 ENST00000253107.7 ENST00000556468.1 ENST00000393793.1 |
PPAN-P2RY11 PPAN |
PPAN-P2RY11 readthrough peter pan homolog (Drosophila) |
| chr14_-_24615523 | 0.25 |
ENST00000559056.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_+_90142884 | 0.24 |
ENST00000369408.5 ENST00000339746.4 ENST00000447838.2 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr10_+_104154229 | 0.24 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr1_+_33283043 | 0.24 |
ENST00000373476.1 ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP |
S100P binding protein |
| chr6_+_137143694 | 0.24 |
ENST00000367756.4 ENST00000541292.1 ENST00000318471.4 |
PEX7 |
peroxisomal biogenesis factor 7 |
| chr19_-_4338838 | 0.24 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr5_+_612387 | 0.23 |
ENST00000264935.5 ENST00000444221.1 |
CEP72 |
centrosomal protein 72kDa |
| chr2_+_231280954 | 0.23 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
| chr3_+_111717600 | 0.23 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr14_+_106355918 | 0.23 |
ENST00000414005.1 |
AL122127.25 |
AL122127.25 |
| chr19_-_4338783 | 0.23 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chrX_+_68048803 | 0.23 |
ENST00000204961.4 |
EFNB1 |
ephrin-B1 |
| chr1_+_27114418 | 0.22 |
ENST00000078527.4 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr17_+_26698677 | 0.22 |
ENST00000457710.3 |
SARM1 |
sterile alpha and TIR motif containing 1 |
| chr1_-_89458415 | 0.22 |
ENST00000321792.5 ENST00000370491.3 |
RBMXL1 CCBL2 |
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr17_+_8924837 | 0.22 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr1_+_26503894 | 0.22 |
ENST00000361530.6 ENST00000374253.5 |
CNKSR1 |
connector enhancer of kinase suppressor of Ras 1 |
| chr9_-_139948468 | 0.21 |
ENST00000312665.5 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr2_-_99771373 | 0.21 |
ENST00000393483.3 |
TSGA10 |
testis specific, 10 |
| chr12_+_125549925 | 0.21 |
ENST00000316519.6 |
AACS |
acetoacetyl-CoA synthetase |
| chr19_+_1450112 | 0.21 |
ENST00000590469.1 ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2 |
adenomatosis polyposis coli 2 |
| chrX_-_1572629 | 0.21 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr2_+_86947296 | 0.21 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr8_+_26149007 | 0.21 |
ENST00000380737.3 ENST00000524169.1 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr8_-_87520849 | 0.20 |
ENST00000430676.2 ENST00000519966.1 |
RMDN1 |
regulator of microtubule dynamics 1 |
| chr14_+_24605361 | 0.20 |
ENST00000206451.6 ENST00000559123.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_+_22235310 | 0.20 |
ENST00000600162.1 |
ZNF257 |
zinc finger protein 257 |
| chr9_-_139948487 | 0.19 |
ENST00000355097.2 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr19_-_14628645 | 0.19 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr15_-_64648273 | 0.19 |
ENST00000607537.1 ENST00000303052.7 ENST00000303032.6 |
CSNK1G1 |
casein kinase 1, gamma 1 |
| chr2_-_197036289 | 0.19 |
ENST00000263955.4 |
STK17B |
serine/threonine kinase 17b |
| chr19_-_49401990 | 0.19 |
ENST00000221399.3 |
TULP2 |
tubby like protein 2 |
| chr1_-_150207017 | 0.19 |
ENST00000369119.3 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr10_+_30722866 | 0.18 |
ENST00000263056.1 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
| chr22_+_35796056 | 0.18 |
ENST00000216122.4 |
MCM5 |
minichromosome maintenance complex component 5 |
| chr7_+_47694842 | 0.18 |
ENST00000408988.2 |
C7orf65 |
chromosome 7 open reading frame 65 |
| chr12_-_54673871 | 0.18 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr12_+_7023491 | 0.17 |
ENST00000541477.1 ENST00000229277.1 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr2_+_242127924 | 0.17 |
ENST00000402530.3 ENST00000274979.8 ENST00000402430.3 |
ANO7 |
anoctamin 7 |
| chr14_+_96152754 | 0.17 |
ENST00000340722.7 |
TCL1B |
T-cell leukemia/lymphoma 1B |
| chr5_-_14871866 | 0.17 |
ENST00000284268.6 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
| chr7_+_87505544 | 0.17 |
ENST00000265728.1 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
| chr5_+_130599735 | 0.17 |
ENST00000503291.1 ENST00000360515.3 ENST00000505065.1 |
CDC42SE2 |
CDC42 small effector 2 |
| chr16_-_15736953 | 0.17 |
ENST00000548025.1 ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430 |
KIAA0430 |
| chr7_-_143059780 | 0.17 |
ENST00000409578.1 ENST00000409346.1 |
FAM131B |
family with sequence similarity 131, member B |
| chr19_-_4831701 | 0.17 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chrX_-_49041242 | 0.17 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr16_-_15736881 | 0.16 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr11_-_3862206 | 0.16 |
ENST00000351018.4 |
RHOG |
ras homolog family member G |
| chr9_-_116139255 | 0.16 |
ENST00000374180.3 |
HDHD3 |
haloacid dehalogenase-like hydrolase domain containing 3 |
| chr10_-_133795416 | 0.16 |
ENST00000540159.1 ENST00000368636.4 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr12_+_120502441 | 0.16 |
ENST00000446727.2 |
CCDC64 |
coiled-coil domain containing 64 |
| chr17_+_7462103 | 0.16 |
ENST00000396545.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_+_56473910 | 0.16 |
ENST00000411731.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr19_-_45927097 | 0.16 |
ENST00000340192.7 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr19_+_33072373 | 0.16 |
ENST00000586035.1 |
PDCD5 |
programmed cell death 5 |
| chr22_+_35796108 | 0.15 |
ENST00000382011.5 ENST00000416905.1 |
MCM5 |
minichromosome maintenance complex component 5 |
| chr2_-_70780770 | 0.15 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr8_+_90770008 | 0.15 |
ENST00000540020.1 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
| chr12_+_57998595 | 0.15 |
ENST00000337737.3 ENST00000548198.1 ENST00000551632.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr8_+_32406179 | 0.15 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr1_+_45965725 | 0.15 |
ENST00000401061.4 |
MMACHC |
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr1_-_89458287 | 0.15 |
ENST00000370485.2 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
| chr3_+_107241783 | 0.15 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr9_+_115913222 | 0.14 |
ENST00000259392.3 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
| chr4_-_860950 | 0.14 |
ENST00000511980.1 ENST00000510799.1 |
GAK |
cyclin G associated kinase |
| chr17_+_16318909 | 0.14 |
ENST00000577397.1 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr19_-_45926739 | 0.14 |
ENST00000589381.1 ENST00000591636.1 ENST00000013807.5 ENST00000592023.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr10_+_22605374 | 0.14 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr20_-_50808525 | 0.14 |
ENST00000216923.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr8_+_128427857 | 0.13 |
ENST00000391675.1 |
POU5F1B |
POU class 5 homeobox 1B |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 4.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 4.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.3 | 2.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 3.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.3 | 2.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 1.6 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.3 | 3.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 0.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 2.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.7 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.1 | 1.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 3.5 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.5 | 1.6 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.4 | 2.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 2.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 3.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.4 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 2.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.9 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.1 | 4.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.3 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0071437 | microvillus membrane(GO:0031528) invadopodium(GO:0071437) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0005923 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 6.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 4.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 3.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1905154 | negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 0.6 | 1.9 | GO:0002316 | follicular B cell differentiation(GO:0002316) activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.5 | 1.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 1.6 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.3 | 3.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 3.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 0.9 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.3 | 0.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 1.0 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.3 | 1.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 2.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.8 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 | 0.6 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.6 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 4.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.5 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.4 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.7 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 1.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.3 | GO:0052552 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.4 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.2 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 1.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.9 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 2.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.2 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) post-embryonic hemopoiesis(GO:0035166) positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.4 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.5 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.0 | 0.6 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 4.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.7 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) cellular response to cobalt ion(GO:0071279) |
| 0.0 | 2.7 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.7 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.6 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 1.3 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.6 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 2.7 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 1.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |