Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
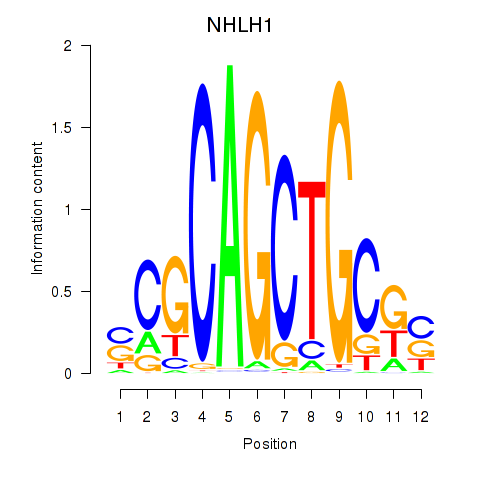
Results for NHLH1
Z-value: 0.55
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | NHLH1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | -0.47 | 6.9e-02 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32022733 | 1.59 |
ENST00000438237.2 ENST00000396556.2 |
OSBPL10 |
oxysterol binding protein-like 10 |
| chr12_-_52887034 | 1.38 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr1_+_20915409 | 0.99 |
ENST00000375071.3 |
CDA |
cytidine deaminase |
| chr12_-_52867569 | 0.92 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr5_+_52776228 | 0.91 |
ENST00000256759.3 |
FST |
follistatin |
| chr20_-_14318248 | 0.87 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr6_+_30848557 | 0.86 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr5_+_52776449 | 0.83 |
ENST00000396947.3 |
FST |
follistatin |
| chr16_+_22825475 | 0.80 |
ENST00000261374.3 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr4_+_75311019 | 0.76 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr4_+_75310851 | 0.72 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr5_-_16936340 | 0.63 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr19_+_38755203 | 0.62 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr5_-_1882858 | 0.62 |
ENST00000511126.1 ENST00000231357.2 |
IRX4 |
iroquois homeobox 4 |
| chr19_+_38755042 | 0.58 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr1_-_237167718 | 0.56 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr5_+_76114758 | 0.52 |
ENST00000514165.1 ENST00000296677.4 |
F2RL1 |
coagulation factor II (thrombin) receptor-like 1 |
| chr20_+_30193083 | 0.52 |
ENST00000376112.3 ENST00000376105.3 |
ID1 |
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr1_-_53387386 | 0.51 |
ENST00000467988.1 ENST00000358358.5 ENST00000371522.4 |
ECHDC2 |
enoyl CoA hydratase domain containing 2 |
| chr2_-_31360887 | 0.49 |
ENST00000420311.2 ENST00000356174.3 ENST00000324589.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_-_209979375 | 0.49 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr11_-_6341844 | 0.48 |
ENST00000303927.3 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr8_-_125740514 | 0.48 |
ENST00000325064.5 ENST00000518547.1 |
MTSS1 |
metastasis suppressor 1 |
| chr1_-_85930823 | 0.47 |
ENST00000284031.8 ENST00000539042.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr8_+_104311059 | 0.45 |
ENST00000358755.4 ENST00000523739.1 ENST00000540287.1 |
FZD6 |
frizzled family receptor 6 |
| chr6_-_30712313 | 0.45 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr8_+_86376081 | 0.45 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr3_-_185542761 | 0.44 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr11_-_27494309 | 0.43 |
ENST00000389858.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_-_66725837 | 0.41 |
ENST00000393958.2 ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC |
pyruvate carboxylase |
| chr11_-_27494279 | 0.41 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr13_-_45150392 | 0.39 |
ENST00000501704.2 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr14_-_61748550 | 0.39 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr4_-_102268484 | 0.39 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_+_44036900 | 0.39 |
ENST00000443296.1 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_-_19051103 | 0.39 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr2_+_69001913 | 0.38 |
ENST00000409030.3 ENST00000409220.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr11_-_6341724 | 0.37 |
ENST00000530979.1 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr18_+_3449695 | 0.37 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr10_+_123923105 | 0.37 |
ENST00000368999.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr20_+_44036620 | 0.36 |
ENST00000372710.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_-_65374430 | 0.35 |
ENST00000532507.1 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
| chr5_+_102201722 | 0.34 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_53491220 | 0.34 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr4_-_102268628 | 0.33 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_45187843 | 0.33 |
ENST00000296129.1 ENST00000425231.2 |
CDCP1 |
CUB domain containing protein 1 |
| chr10_+_123922941 | 0.33 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr18_+_3449821 | 0.33 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_-_54423529 | 0.32 |
ENST00000245451.4 ENST00000559087.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr6_+_30851840 | 0.32 |
ENST00000511510.1 ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_27278304 | 0.32 |
ENST00000577226.1 |
PHF12 |
PHD finger protein 12 |
| chr10_+_123923205 | 0.32 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_68771128 | 0.31 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr17_+_39411636 | 0.31 |
ENST00000394008.1 |
KRTAP9-9 |
keratin associated protein 9-9 |
| chrX_+_9754461 | 0.31 |
ENST00000380913.3 |
SHROOM2 |
shroom family member 2 |
| chr11_+_394196 | 0.31 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr5_+_102201687 | 0.30 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr10_-_103347883 | 0.30 |
ENST00000339310.3 ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL |
polymerase (DNA directed), lambda |
| chr5_+_102201509 | 0.30 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_41216492 | 0.29 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_+_150151752 | 0.28 |
ENST00000325307.7 |
HMGB3 |
high mobility group box 3 |
| chr4_-_41216473 | 0.28 |
ENST00000513140.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_-_6662919 | 0.27 |
ENST00000377658.4 ENST00000377663.3 |
KLHL21 |
kelch-like family member 21 |
| chrX_+_110187513 | 0.27 |
ENST00000446737.1 ENST00000425146.1 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chrX_+_107069063 | 0.26 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr2_-_235405168 | 0.26 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr1_-_225840747 | 0.25 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr11_-_35441524 | 0.25 |
ENST00000395750.1 ENST00000449068.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_27170352 | 0.25 |
ENST00000428284.2 ENST00000360046.5 |
HOXA4 |
homeobox A4 |
| chr1_+_13910479 | 0.24 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr12_+_57984965 | 0.23 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr5_-_95297534 | 0.22 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr11_+_67070919 | 0.22 |
ENST00000308127.4 ENST00000308298.7 |
SSH3 |
slingshot protein phosphatase 3 |
| chr3_-_99833333 | 0.22 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr16_-_29910365 | 0.22 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr18_+_3448455 | 0.21 |
ENST00000549780.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr11_-_35441597 | 0.21 |
ENST00000395753.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr12_-_118541743 | 0.21 |
ENST00000359236.5 |
VSIG10 |
V-set and immunoglobulin domain containing 10 |
| chr21_-_46293644 | 0.20 |
ENST00000330938.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr3_-_119813264 | 0.20 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr6_+_31895254 | 0.20 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr1_-_182360918 | 0.20 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr4_-_139163491 | 0.20 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_+_13910194 | 0.20 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr1_+_60280458 | 0.20 |
ENST00000455990.1 ENST00000371208.3 |
HOOK1 |
hook microtubule-tethering protein 1 |
| chr2_-_178483694 | 0.20 |
ENST00000355689.5 |
TTC30A |
tetratricopeptide repeat domain 30A |
| chr4_-_41216619 | 0.20 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr11_+_71249071 | 0.19 |
ENST00000398534.3 |
KRTAP5-8 |
keratin associated protein 5-8 |
| chr7_-_27169801 | 0.19 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr21_-_46293586 | 0.19 |
ENST00000445724.2 ENST00000397887.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr10_-_35104185 | 0.19 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr1_-_19536744 | 0.19 |
ENST00000375267.2 ENST00000375217.2 ENST00000375226.2 ENST00000375254.3 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
| chr1_+_90287480 | 0.19 |
ENST00000394593.3 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr19_+_45281118 | 0.19 |
ENST00000270279.3 ENST00000341505.4 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr11_+_64948665 | 0.18 |
ENST00000533820.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr6_+_31895467 | 0.18 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr14_-_104313824 | 0.18 |
ENST00000553739.1 ENST00000202556.9 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
| chr4_+_154074217 | 0.18 |
ENST00000437508.2 |
TRIM2 |
tripartite motif containing 2 |
| chr2_+_236578248 | 0.18 |
ENST00000409538.1 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr2_+_70485220 | 0.18 |
ENST00000433351.2 ENST00000264441.5 |
PCYOX1 |
prenylcysteine oxidase 1 |
| chr6_+_31895480 | 0.18 |
ENST00000418949.2 ENST00000383177.3 ENST00000477310.1 |
C2 CFB |
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_-_182360498 | 0.18 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chrX_-_84634737 | 0.18 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr19_-_47164386 | 0.17 |
ENST00000391916.2 ENST00000410105.2 |
DACT3 |
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_80657292 | 0.17 |
ENST00000369816.4 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
| chr2_-_73340146 | 0.17 |
ENST00000258098.6 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
| chr15_+_25200074 | 0.17 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr1_+_955448 | 0.17 |
ENST00000379370.2 |
AGRN |
agrin |
| chr14_+_62162258 | 0.16 |
ENST00000337138.4 ENST00000394997.1 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr1_-_182361327 | 0.16 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr14_-_24551195 | 0.16 |
ENST00000560550.1 |
NRL |
neural retina leucine zipper |
| chr5_-_95297678 | 0.16 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr20_+_34043085 | 0.16 |
ENST00000397527.1 ENST00000342580.4 |
CEP250 |
centrosomal protein 250kDa |
| chr7_-_11871815 | 0.16 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr4_-_54930790 | 0.16 |
ENST00000263921.3 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
| chr17_-_39191107 | 0.15 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr18_-_30050395 | 0.15 |
ENST00000269209.6 ENST00000399218.4 |
GAREM |
GRB2 associated, regulator of MAPK1 |
| chr8_-_144897549 | 0.15 |
ENST00000356994.2 ENST00000320476.3 |
SCRIB |
scribbled planar cell polarity protein |
| chr8_-_144897138 | 0.15 |
ENST00000377533.3 |
SCRIB |
scribbled planar cell polarity protein |
| chr16_-_90085824 | 0.15 |
ENST00000002501.6 |
DBNDD1 |
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr16_+_1756162 | 0.15 |
ENST00000250894.4 ENST00000356010.5 |
MAPK8IP3 |
mitogen-activated protein kinase 8 interacting protein 3 |
| chr13_-_96296944 | 0.15 |
ENST00000361396.2 ENST00000376829.2 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
| chr7_-_148725733 | 0.15 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr7_+_766320 | 0.15 |
ENST00000297440.6 ENST00000313147.5 |
HEATR2 |
HEAT repeat containing 2 |
| chr9_+_114393634 | 0.15 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr19_+_35521572 | 0.15 |
ENST00000262631.5 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
| chrX_+_150151824 | 0.15 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr12_-_123752624 | 0.14 |
ENST00000542174.1 ENST00000535796.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr14_-_24551137 | 0.14 |
ENST00000396995.1 |
NRL |
neural retina leucine zipper |
| chr19_+_17666403 | 0.14 |
ENST00000252599.4 |
COLGALT1 |
collagen beta(1-O)galactosyltransferase 1 |
| chr1_-_38273840 | 0.14 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr1_+_15272271 | 0.14 |
ENST00000400797.3 |
KAZN |
kazrin, periplakin interacting protein |
| chr1_+_90286562 | 0.14 |
ENST00000525774.1 ENST00000337338.5 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr7_-_123389104 | 0.14 |
ENST00000223023.4 |
WASL |
Wiskott-Aldrich syndrome-like |
| chr16_-_67694129 | 0.14 |
ENST00000602320.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr20_-_33680635 | 0.14 |
ENST00000252015.2 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr20_-_33680588 | 0.13 |
ENST00000451813.2 ENST00000432634.2 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr3_+_142442841 | 0.13 |
ENST00000476941.1 ENST00000273482.6 |
TRPC1 |
transient receptor potential cation channel, subfamily C, member 1 |
| chr4_+_30723003 | 0.13 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chr6_+_43395272 | 0.13 |
ENST00000372530.4 |
ABCC10 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr20_+_47538357 | 0.13 |
ENST00000371917.4 |
ARFGEF2 |
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr15_-_52472078 | 0.13 |
ENST00000396335.4 ENST00000560116.1 ENST00000358784.7 |
GNB5 |
guanine nucleotide binding protein (G protein), beta 5 |
| chr8_+_24772455 | 0.13 |
ENST00000433454.2 |
NEFM |
neurofilament, medium polypeptide |
| chr17_+_28705921 | 0.13 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr14_-_107211459 | 0.13 |
ENST00000390636.2 |
IGHV3-73 |
immunoglobulin heavy variable 3-73 |
| chr4_+_3768075 | 0.13 |
ENST00000509482.1 ENST00000330055.5 |
ADRA2C |
adrenoceptor alpha 2C |
| chr6_+_43543864 | 0.12 |
ENST00000372236.4 ENST00000535400.1 |
POLH |
polymerase (DNA directed), eta |
| chr13_+_28194873 | 0.12 |
ENST00000302979.3 |
POLR1D |
polymerase (RNA) I polypeptide D, 16kDa |
| chr7_+_99613212 | 0.12 |
ENST00000426572.1 ENST00000535170.1 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr15_+_25200108 | 0.12 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr19_+_16186903 | 0.12 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chr16_+_67465016 | 0.12 |
ENST00000326152.5 |
HSD11B2 |
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr7_-_44924939 | 0.12 |
ENST00000395699.2 |
PURB |
purine-rich element binding protein B |
| chr2_+_27274506 | 0.12 |
ENST00000451003.1 ENST00000323064.8 ENST00000360131.4 |
AGBL5 |
ATP/GTP binding protein-like 5 |
| chr11_-_34379546 | 0.12 |
ENST00000435224.2 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr17_+_55162453 | 0.12 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr7_+_130131907 | 0.12 |
ENST00000223215.4 ENST00000437945.1 |
MEST |
mesoderm specific transcript |
| chr17_-_66453562 | 0.12 |
ENST00000262139.5 ENST00000546360.1 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
| chr14_+_24583836 | 0.11 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr1_-_21671968 | 0.11 |
ENST00000415912.2 |
ECE1 |
endothelin converting enzyme 1 |
| chr4_-_89744365 | 0.11 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr22_+_32340481 | 0.11 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr19_+_48876300 | 0.11 |
ENST00000600863.1 ENST00000601610.1 ENST00000595322.1 |
SYNGR4 |
synaptogyrin 4 |
| chr20_+_34042962 | 0.11 |
ENST00000446710.1 ENST00000420564.1 |
CEP250 |
centrosomal protein 250kDa |
| chr1_+_202317815 | 0.11 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr9_+_125703282 | 0.11 |
ENST00000373647.4 ENST00000402311.1 |
RABGAP1 |
RAB GTPase activating protein 1 |
| chr14_-_24911868 | 0.11 |
ENST00000554698.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr8_+_42995548 | 0.11 |
ENST00000458501.2 ENST00000379644.4 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
| chr6_+_43543942 | 0.11 |
ENST00000372226.1 ENST00000443535.1 |
POLH |
polymerase (DNA directed), eta |
| chr6_-_31681839 | 0.11 |
ENST00000409239.1 ENST00000461287.1 |
LY6G6E XXbac-BPG32J3.20 |
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr22_+_32340447 | 0.11 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr12_+_122064398 | 0.11 |
ENST00000330079.7 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr13_-_72440901 | 0.11 |
ENST00000359684.2 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr4_-_122854193 | 0.10 |
ENST00000513531.1 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr2_+_233562015 | 0.10 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr3_+_5020801 | 0.10 |
ENST00000256495.3 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
| chr16_-_82203780 | 0.10 |
ENST00000563504.1 ENST00000569021.1 ENST00000258169.4 |
MPHOSPH6 |
M-phase phosphoprotein 6 |
| chr12_-_110318226 | 0.10 |
ENST00000544393.1 |
GLTP |
glycolipid transfer protein |
| chr8_-_67579418 | 0.10 |
ENST00000310421.4 |
VCPIP1 |
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr1_-_27693349 | 0.10 |
ENST00000374040.3 ENST00000357582.2 ENST00000493901.1 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
| chr14_-_24584138 | 0.10 |
ENST00000558280.1 ENST00000561028.1 |
NRL |
neural retina leucine zipper |
| chr3_+_124303539 | 0.10 |
ENST00000428018.2 |
KALRN |
kalirin, RhoGEF kinase |
| chr1_+_24969755 | 0.10 |
ENST00000447431.2 ENST00000374389.4 |
SRRM1 |
serine/arginine repetitive matrix 1 |
| chr4_-_89744314 | 0.10 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr7_-_128045984 | 0.10 |
ENST00000470772.1 ENST00000480861.1 ENST00000496200.1 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chrX_+_47004599 | 0.10 |
ENST00000329236.7 |
RBM10 |
RNA binding motif protein 10 |
| chr1_-_3713042 | 0.10 |
ENST00000378251.1 |
LRRC47 |
leucine rich repeat containing 47 |
| chr2_+_231902193 | 0.10 |
ENST00000373640.4 |
C2orf72 |
chromosome 2 open reading frame 72 |
| chr5_-_94620239 | 0.10 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr6_-_82462425 | 0.10 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr1_+_156105878 | 0.09 |
ENST00000508500.1 |
LMNA |
lamin A/C |
| chr14_-_24911971 | 0.09 |
ENST00000555365.1 ENST00000399395.3 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr17_-_79139817 | 0.09 |
ENST00000326724.4 |
AATK |
apoptosis-associated tyrosine kinase |
| chr16_+_67562702 | 0.09 |
ENST00000379312.3 ENST00000042381.4 ENST00000540839.3 |
FAM65A |
family with sequence similarity 65, member A |
| chr1_+_78511586 | 0.09 |
ENST00000370759.3 |
GIPC2 |
GIPC PDZ domain containing family, member 2 |
| chr1_+_3388181 | 0.09 |
ENST00000418137.1 ENST00000413250.2 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr19_+_39989535 | 0.09 |
ENST00000356433.5 |
DLL3 |
delta-like 3 (Drosophila) |
| chr20_+_8112824 | 0.09 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr21_+_34697258 | 0.09 |
ENST00000442071.1 ENST00000442357.2 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
| chr7_-_32110451 | 0.09 |
ENST00000396191.1 ENST00000396182.2 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 1.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.3 | 1.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.7 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.9 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.2 | 1.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.8 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 1.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 0.5 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 0.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.2 | 0.2 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.4 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.5 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.3 | GO:0048390 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.3 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.9 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 1.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.5 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 1.0 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.4 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.5 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.9 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.4 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.4 | GO:0008301 | four-way junction DNA binding(GO:0000400) DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |