Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NKX1-1
Z-value: 0.34
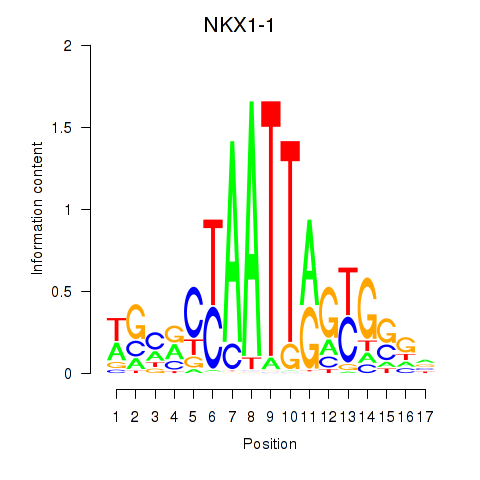
Transcription factors associated with NKX1-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-1
|
ENSG00000235608.1 | NKX1-1 |
Activity profile of NKX1-1 motif
Sorted Z-values of NKX1-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_8822113 | 1.22 |
ENST00000396290.1 ENST00000331129.3 |
ID2 |
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr1_-_221915418 | 0.56 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr7_-_86849883 | 0.53 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr7_+_26331541 | 0.52 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr3_-_105588231 | 0.43 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_+_128554081 | 0.39 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr8_+_26371763 | 0.32 |
ENST00000521913.1 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr1_+_174843548 | 0.25 |
ENST00000478442.1 ENST00000465412.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr2_-_239197238 | 0.22 |
ENST00000254657.3 |
PER2 |
period circadian clock 2 |
| chr18_-_33709268 | 0.20 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr3_+_57261743 | 0.20 |
ENST00000288266.3 |
APPL1 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr17_-_61777459 | 0.19 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr2_-_239197201 | 0.18 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr14_-_24711865 | 0.17 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_-_105587879 | 0.17 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr14_-_24711806 | 0.16 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr17_-_8113886 | 0.16 |
ENST00000577833.1 ENST00000534871.1 ENST00000583915.1 ENST00000316199.6 ENST00000581511.1 ENST00000585124.1 |
AURKB |
aurora kinase B |
| chr12_-_6961050 | 0.15 |
ENST00000538862.2 |
CDCA3 |
cell division cycle associated 3 |
| chr12_+_7014126 | 0.15 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr12_+_7014064 | 0.13 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr17_-_45266542 | 0.13 |
ENST00000531206.1 ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27 |
cell division cycle 27 |
| chr8_+_98656693 | 0.13 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr7_+_50348268 | 0.13 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr12_+_7013897 | 0.12 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr8_+_98656336 | 0.11 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr15_-_78526855 | 0.11 |
ENST00000541759.1 ENST00000558130.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_23851928 | 0.10 |
ENST00000467766.1 ENST00000424381.1 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr1_+_46972668 | 0.09 |
ENST00000371956.4 ENST00000360032.3 |
DMBX1 |
diencephalon/mesencephalon homeobox 1 |
| chr17_-_44657017 | 0.08 |
ENST00000573185.1 ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A |
ADP-ribosylation factor-like 17A |
| chr17_-_44439084 | 0.08 |
ENST00000575960.1 ENST00000575698.1 ENST00000571246.1 ENST00000434041.2 ENST00000570618.1 ENST00000450673.3 |
ARL17B |
ADP-ribosylation factor-like 17B |
| chr14_-_24711470 | 0.08 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_-_111741994 | 0.07 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr11_-_71823715 | 0.07 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr11_-_62494821 | 0.07 |
ENST00000301785.5 |
HNRNPUL2 |
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr11_-_71823796 | 0.07 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr1_-_51796226 | 0.06 |
ENST00000451380.1 ENST00000371747.3 ENST00000439482.1 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr1_-_108507631 | 0.06 |
ENST00000527011.1 ENST00000370056.4 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr19_-_6424783 | 0.06 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr12_+_102513950 | 0.06 |
ENST00000378128.3 ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP |
PARP1 binding protein |
| chr19_+_36249057 | 0.06 |
ENST00000301165.5 ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55 |
chromosome 19 open reading frame 55 |
| chr4_-_103682145 | 0.06 |
ENST00000226578.4 |
MANBA |
mannosidase, beta A, lysosomal |
| chr4_-_8873531 | 0.05 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chr14_+_32798462 | 0.05 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr1_+_62439037 | 0.05 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr9_+_139780942 | 0.05 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr19_+_45504688 | 0.05 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_+_187371440 | 0.04 |
ENST00000445547.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr17_-_8055747 | 0.04 |
ENST00000317276.4 ENST00000581703.1 |
PER1 |
period circadian clock 1 |
| chr17_-_27418537 | 0.04 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr1_-_51796987 | 0.04 |
ENST00000262676.5 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr10_-_1095050 | 0.03 |
ENST00000381344.3 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
| chr22_-_39151463 | 0.03 |
ENST00000405510.1 ENST00000433561.1 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
| chr12_-_23737534 | 0.03 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr12_+_2921788 | 0.03 |
ENST00000228799.2 ENST00000419778.2 ENST00000542548.1 |
ITFG2 |
integrin alpha FG-GAP repeat containing 2 |
| chr6_-_82957433 | 0.03 |
ENST00000306270.7 |
IBTK |
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr10_+_120789223 | 0.03 |
ENST00000425699.1 |
NANOS1 |
nanos homolog 1 (Drosophila) |
| chr12_-_91451758 | 0.03 |
ENST00000266719.3 |
KERA |
keratocan |
| chr15_+_64386261 | 0.03 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chrX_-_47509887 | 0.02 |
ENST00000247161.3 ENST00000592066.1 ENST00000376983.3 |
ELK1 |
ELK1, member of ETS oncogene family |
| chr3_+_186560476 | 0.02 |
ENST00000320741.2 ENST00000444204.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560462 | 0.02 |
ENST00000412955.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr13_-_36050819 | 0.02 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr17_-_40337470 | 0.02 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr4_-_66536196 | 0.02 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr19_-_8070474 | 0.01 |
ENST00000407627.2 ENST00000593807.1 |
ELAVL1 |
ELAV like RNA binding protein 1 |
| chr12_+_28410128 | 0.01 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr7_+_144052381 | 0.01 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr20_-_50722183 | 0.01 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr4_-_66536057 | 0.01 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chrX_-_153363125 | 0.01 |
ENST00000407218.1 ENST00000453960.2 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chrX_+_55026763 | 0.01 |
ENST00000374987.3 |
APEX2 |
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr17_-_10372875 | 0.01 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr11_+_72983246 | 0.01 |
ENST00000393590.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr1_-_190446759 | 0.01 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr15_-_78526942 | 0.00 |
ENST00000258873.4 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |