Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NKX2-1
Z-value: 0.75
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.13 | NKX2-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg19_v2_chr14_-_36989336_36989354 | -0.20 | 4.5e-01 | Click! |
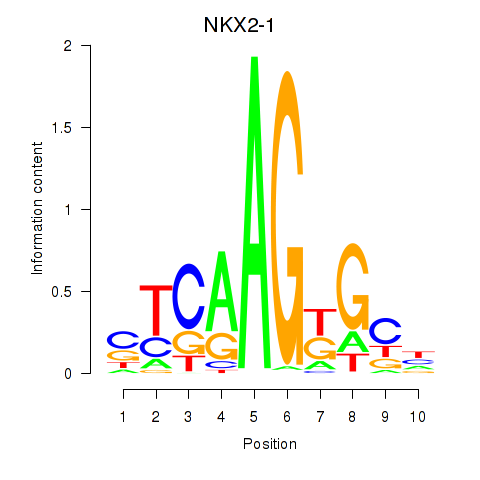
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
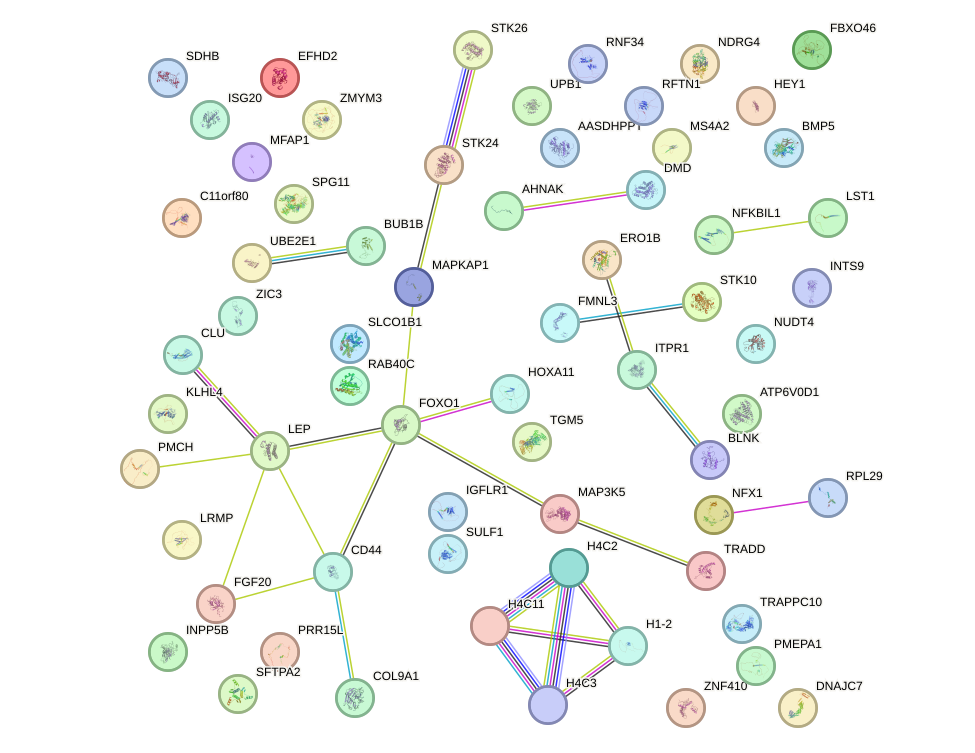
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_60223225 | 3.14 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_60223312 | 3.14 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr10_-_98031310 | 2.05 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr10_-_98031265 | 1.98 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr12_-_21810765 | 1.90 |
ENST00000450584.1 ENST00000350669.1 |
LDHB |
lactate dehydrogenase B |
| chr12_-_21810726 | 1.90 |
ENST00000396076.1 |
LDHB |
lactate dehydrogenase B |
| chr3_-_16524357 | 1.53 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr12_+_25205666 | 1.46 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr12_+_25205568 | 1.44 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr6_-_137113604 | 1.30 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr12_+_25205446 | 1.22 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr15_+_89182178 | 1.17 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr1_-_25291475 | 1.14 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr15_+_89182156 | 1.07 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89181974 | 1.03 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr11_+_35160709 | 0.87 |
ENST00000415148.2 ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44 |
CD44 molecule (Indian blood group) |
| chr1_-_38397384 | 0.80 |
ENST00000373027.1 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
| chr5_-_171615315 | 0.80 |
ENST00000176763.5 |
STK10 |
serine/threonine kinase 10 |
| chr6_-_26056695 | 0.79 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr13_-_41240717 | 0.75 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr12_-_719573 | 0.74 |
ENST00000397265.3 |
NINJ2 |
ninjurin 2 |
| chrX_+_131157322 | 0.70 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_+_131157290 | 0.69 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr8_-_80680078 | 0.66 |
ENST00000337919.5 ENST00000354724.3 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr19_-_36231437 | 0.56 |
ENST00000591748.1 |
IGFLR1 |
IGF-like family receptor 1 |
| chr19_+_1065922 | 0.55 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr1_-_236445251 | 0.51 |
ENST00000354619.5 ENST00000327333.8 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
| chrX_-_32173579 | 0.51 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr3_+_4535025 | 0.51 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr12_+_93772402 | 0.51 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr8_-_28747424 | 0.48 |
ENST00000523436.1 ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9 |
integrator complex subunit 9 |
| chr15_-_44116873 | 0.48 |
ENST00000267812.3 |
MFAP1 |
microfibrillar-associated protein 1 |
| chr19_-_46234119 | 0.48 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr17_-_76778339 | 0.45 |
ENST00000591455.1 ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1 |
cytohesin 1 |
| chr16_-_67190152 | 0.43 |
ENST00000486556.1 |
TRADD |
TNFRSF1A-associated via death domain |
| chr15_-_43559055 | 0.41 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr8_-_16859690 | 0.38 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr12_+_121837905 | 0.37 |
ENST00000392465.3 ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
| chr1_-_17380630 | 0.35 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr12_+_93772326 | 0.35 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr3_+_23847432 | 0.34 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr9_+_33290491 | 0.34 |
ENST00000379540.3 ENST00000379521.4 ENST00000318524.6 |
NFX1 |
nuclear transcription factor, X-box binding 1 |
| chr9_-_128246769 | 0.32 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr15_-_44955842 | 0.31 |
ENST00000427534.2 ENST00000559193.1 ENST00000261866.7 ENST00000535302.2 ENST00000558319.1 |
SPG11 |
spastic paraplegia 11 (autosomal recessive) |
| chr22_+_24891210 | 0.30 |
ENST00000382760.2 |
UPB1 |
ureidopropionase, beta |
| chrX_+_136648297 | 0.29 |
ENST00000287538.5 |
ZIC3 |
Zic family member 3 |
| chr11_+_105948216 | 0.29 |
ENST00000278618.4 |
AASDHPPT |
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr19_+_36235964 | 0.29 |
ENST00000587708.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr16_+_640055 | 0.27 |
ENST00000568586.1 ENST00000538492.1 ENST00000248139.3 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr19_+_48958766 | 0.27 |
ENST00000342291.2 |
KCNJ14 |
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr21_+_45432174 | 0.26 |
ENST00000380221.3 ENST00000291574.4 |
TRAPPC10 |
trafficking protein particle complex 10 |
| chr18_+_33877654 | 0.26 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr7_-_27224795 | 0.24 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr18_+_42260861 | 0.24 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr1_+_15736359 | 0.23 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr20_-_56285595 | 0.22 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr8_+_70404996 | 0.21 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr16_+_1359138 | 0.21 |
ENST00000325437.5 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chr6_+_31554779 | 0.20 |
ENST00000376090.2 |
LST1 |
leukocyte specific transcript 1 |
| chr3_+_4535155 | 0.19 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chrX_-_110655306 | 0.19 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr7_-_27224842 | 0.18 |
ENST00000517402.1 |
HOXA11 |
homeobox A11 |
| chr1_+_206643787 | 0.17 |
ENST00000367120.3 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr5_+_134094461 | 0.16 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr16_-_67493110 | 0.15 |
ENST00000602876.1 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr6_-_71012773 | 0.14 |
ENST00000370496.3 ENST00000357250.6 |
COL9A1 |
collagen, type IX, alpha 1 |
| chr1_+_206643806 | 0.14 |
ENST00000537984.1 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr12_-_102591604 | 0.13 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr14_+_74353320 | 0.13 |
ENST00000540593.1 ENST00000555730.1 |
ZNF410 |
zinc finger protein 410 |
| chr16_+_58533951 | 0.13 |
ENST00000566192.1 ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4 |
NDRG family member 4 |
| chr17_-_40169429 | 0.13 |
ENST00000316603.7 ENST00000588641.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr19_+_24009879 | 0.12 |
ENST00000354585.4 |
RPSAP58 |
ribosomal protein SA pseudogene 58 |
| chr8_+_94752349 | 0.12 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr7_+_127881325 | 0.12 |
ENST00000308868.4 |
LEP |
leptin |
| chr6_-_27841289 | 0.12 |
ENST00000355981.2 |
HIST1H4L |
histone cluster 1, H4l |
| chr19_+_48972265 | 0.12 |
ENST00000452733.2 |
CYTH2 |
cytohesin 2 |
| chr10_-_81320151 | 0.11 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chr1_+_24019099 | 0.11 |
ENST00000443624.1 ENST00000458455.1 |
RPL11 |
ribosomal protein L11 |
| chr3_-_52029830 | 0.11 |
ENST00000492277.1 ENST00000475248.1 ENST00000479017.1 ENST00000495383.1 ENST00000466397.1 ENST00000481629.1 |
RPL29 |
ribosomal protein L29 |
| chr6_-_55740352 | 0.11 |
ENST00000370830.3 |
BMP5 |
bone morphogenetic protein 5 |
| chr15_+_40453204 | 0.11 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr21_+_45773515 | 0.10 |
ENST00000397932.2 ENST00000300481.9 |
TRPM2 |
transient receptor potential cation channel, subfamily M, member 2 |
| chr19_+_48972459 | 0.10 |
ENST00000427476.1 |
CYTH2 |
cytohesin 2 |
| chr3_-_52029958 | 0.09 |
ENST00000294189.6 |
RPL29 |
ribosomal protein L29 |
| chr17_-_46035187 | 0.09 |
ENST00000300557.2 |
PRR15L |
proline rich 15-like |
| chr19_-_10679644 | 0.09 |
ENST00000393599.2 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chrX_+_153170189 | 0.09 |
ENST00000358927.2 |
AVPR2 |
arginine vasopressin receptor 2 |
| chrX_+_86772707 | 0.08 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chr11_+_66512089 | 0.08 |
ENST00000524551.1 ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80 |
chromosome 11 open reading frame 80 |
| chrX_-_70474499 | 0.08 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr11_+_66512303 | 0.08 |
ENST00000532565.2 |
C11orf80 |
chromosome 11 open reading frame 80 |
| chr12_+_60058458 | 0.07 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_62313090 | 0.07 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr17_-_10452929 | 0.06 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr6_-_55739542 | 0.06 |
ENST00000446683.2 |
BMP5 |
bone morphogenetic protein 5 |
| chr2_+_220379052 | 0.06 |
ENST00000347842.3 ENST00000358078.4 |
ASIC4 |
acid-sensing (proton-gated) ion channel family member 4 |
| chr1_-_92952433 | 0.06 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr12_+_50478977 | 0.05 |
ENST00000381513.4 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_-_228604328 | 0.05 |
ENST00000355586.4 ENST00000366698.2 ENST00000520264.1 ENST00000479800.1 ENST00000295033.3 |
TRIM17 |
tripartite motif containing 17 |
| chr17_-_79623597 | 0.04 |
ENST00000574024.1 ENST00000331056.5 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr14_+_67708137 | 0.04 |
ENST00000556345.1 ENST00000555925.1 ENST00000557783.1 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr3_-_114790179 | 0.04 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_140345820 | 0.04 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr19_+_38924316 | 0.04 |
ENST00000355481.4 ENST00000360985.3 ENST00000359596.3 |
RYR1 |
ryanodine receptor 1 (skeletal) |
| chr9_-_97356075 | 0.04 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr14_+_81421861 | 0.03 |
ENST00000298171.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr14_+_81421921 | 0.03 |
ENST00000554263.1 ENST00000554435.1 |
TSHR |
thyroid stimulating hormone receptor |
| chrX_-_83442915 | 0.03 |
ENST00000262752.2 ENST00000543399.1 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chrX_+_153170455 | 0.03 |
ENST00000430697.1 ENST00000337474.5 ENST00000370049.1 |
AVPR2 |
arginine vasopressin receptor 2 |
| chr6_-_160679905 | 0.03 |
ENST00000366953.3 |
SLC22A2 |
solute carrier family 22 (organic cation transporter), member 2 |
| chr18_-_24283586 | 0.03 |
ENST00000579458.1 |
U3 |
Small nucleolar RNA U3 |
| chr8_+_22019168 | 0.02 |
ENST00000318561.3 ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC |
surfactant protein C |
| chr14_-_58893832 | 0.02 |
ENST00000556007.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_106068065 | 0.02 |
ENST00000390541.2 |
IGHE |
immunoglobulin heavy constant epsilon |
| chr2_+_99797542 | 0.02 |
ENST00000338148.3 ENST00000512183.2 |
MRPL30 C2orf15 |
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr16_+_640201 | 0.02 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr2_-_85895295 | 0.01 |
ENST00000428225.1 ENST00000519937.2 |
SFTPB |
surfactant protein B |
| chr12_-_67197760 | 0.01 |
ENST00000539540.1 ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr11_+_22688150 | 0.01 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chrX_-_134478012 | 0.00 |
ENST00000370766.3 |
ZNF75D |
zinc finger protein 75D |
| chr8_-_144679296 | 0.00 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chrX_+_78003204 | 0.00 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr19_+_55014085 | 0.00 |
ENST00000351841.2 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
| chr17_+_76210267 | 0.00 |
ENST00000301633.4 ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 3.8 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 4.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 6.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.9 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.2 | 0.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.3 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.8 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 3.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.5 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.2 | 1.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 0.9 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 1.3 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.4 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.2 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 6.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:2000366 | negative regulation of glucagon secretion(GO:0070093) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 3.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 4.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.8 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.4 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 4.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 3.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 4.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 3.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |