Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
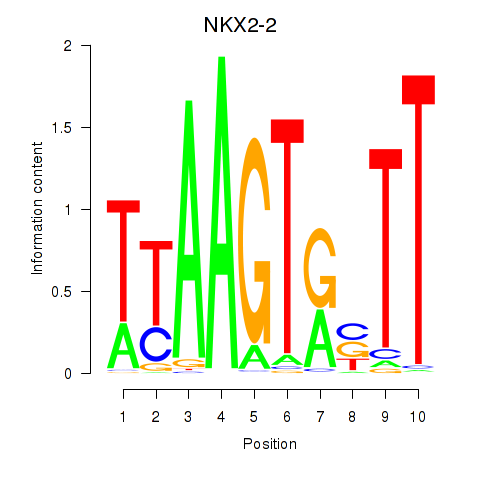
Results for NKX2-2
Z-value: 0.47
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NKX2-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-2 | hg19_v2_chr20_-_21494654_21494678 | -0.07 | 8.0e-01 | Click! |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_55888767 | 0.64 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_74269956 | 0.62 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr12_-_54689532 | 0.60 |
ENST00000540264.2 ENST00000312156.4 |
NFE2 |
nuclear factor, erythroid 2 |
| chr1_-_227505289 | 0.52 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_+_152953505 | 0.49 |
ENST00000253122.5 |
SLC6A8 |
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr16_+_30212378 | 0.47 |
ENST00000569485.1 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr16_+_30212050 | 0.46 |
ENST00000563322.1 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr15_+_58702742 | 0.46 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr5_+_135394840 | 0.46 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr17_+_52978107 | 0.43 |
ENST00000445275.2 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chrX_+_115567767 | 0.40 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr13_-_67802549 | 0.37 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr8_+_97597148 | 0.35 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr4_-_111563076 | 0.34 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr17_+_53342311 | 0.33 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr15_-_56209306 | 0.32 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr6_+_158733692 | 0.32 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr6_+_32121789 | 0.32 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr17_+_74372662 | 0.31 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr9_-_99382065 | 0.30 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr2_+_170655322 | 0.29 |
ENST00000260956.4 ENST00000417292.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr1_+_54359854 | 0.29 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr6_+_32121218 | 0.27 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr1_-_202936394 | 0.25 |
ENST00000367249.4 |
CYB5R1 |
cytochrome b5 reductase 1 |
| chrX_-_154033661 | 0.24 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr12_-_28124903 | 0.24 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr5_+_108083517 | 0.24 |
ENST00000281092.4 ENST00000536402.1 |
FER |
fer (fps/fes related) tyrosine kinase |
| chr5_+_82767487 | 0.24 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr8_+_77593448 | 0.23 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr4_+_155484155 | 0.23 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr3_-_114790179 | 0.23 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_+_88720698 | 0.22 |
ENST00000226284.5 |
IBSP |
integrin-binding sialoprotein |
| chr2_+_17721230 | 0.21 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chr5_+_82767583 | 0.21 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr12_-_8803128 | 0.21 |
ENST00000543467.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr13_-_36429763 | 0.21 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr4_+_80584903 | 0.20 |
ENST00000506460.1 |
RP11-452C8.1 |
RP11-452C8.1 |
| chr16_-_20556492 | 0.20 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr3_-_196911002 | 0.19 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr14_+_39734482 | 0.18 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr11_-_128894053 | 0.18 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr17_+_61086917 | 0.18 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr15_+_65843130 | 0.18 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr8_-_49833978 | 0.17 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr9_+_131447342 | 0.17 |
ENST00000409104.3 |
SET |
SET nuclear oncogene |
| chr11_-_72432950 | 0.17 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr8_-_141810634 | 0.17 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr7_+_2394445 | 0.16 |
ENST00000360876.4 ENST00000413917.1 ENST00000397011.2 |
EIF3B |
eukaryotic translation initiation factor 3, subunit B |
| chr17_+_75315534 | 0.16 |
ENST00000590294.1 ENST00000329047.8 |
SEPT9 |
septin 9 |
| chr8_+_77593474 | 0.16 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr10_+_13203543 | 0.16 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr13_+_50656307 | 0.16 |
ENST00000378180.4 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr8_-_49834299 | 0.16 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr3_-_33686743 | 0.16 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr11_-_77850629 | 0.16 |
ENST00000376156.3 ENST00000525870.1 ENST00000530454.1 ENST00000525755.1 ENST00000527099.1 ENST00000525761.1 ENST00000299626.5 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
| chr1_-_153363452 | 0.16 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr11_+_9406169 | 0.15 |
ENST00000379719.3 ENST00000527431.1 |
IPO7 |
importin 7 |
| chr13_+_50570019 | 0.15 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr19_-_14889349 | 0.15 |
ENST00000315576.3 ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr4_+_155484103 | 0.15 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr17_+_66521936 | 0.14 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_-_70474910 | 0.14 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr17_+_48823896 | 0.14 |
ENST00000511974.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr22_-_36220420 | 0.14 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_+_159848854 | 0.14 |
ENST00000517480.1 ENST00000520452.1 ENST00000393964.1 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr3_-_170744498 | 0.14 |
ENST00000382808.4 ENST00000314251.3 |
SLC2A2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chrM_+_4431 | 0.13 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr2_+_103035102 | 0.13 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chrX_-_32173579 | 0.13 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr5_+_148206156 | 0.13 |
ENST00000305988.4 |
ADRB2 |
adrenoceptor beta 2, surface |
| chr2_-_145275228 | 0.12 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_102591604 | 0.12 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr4_+_41937131 | 0.12 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr5_-_78281603 | 0.12 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr8_-_18666360 | 0.12 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr4_-_76957214 | 0.12 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr4_-_10686475 | 0.11 |
ENST00000226951.6 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr9_+_92219919 | 0.11 |
ENST00000252506.6 ENST00000375769.1 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
| chr4_+_95376396 | 0.11 |
ENST00000508216.1 ENST00000514743.1 |
PDLIM5 |
PDZ and LIM domain 5 |
| chrX_+_123095546 | 0.11 |
ENST00000371157.3 ENST00000371145.3 ENST00000371144.3 |
STAG2 |
stromal antigen 2 |
| chr8_-_141774467 | 0.11 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr1_-_244006528 | 0.11 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr1_-_36615065 | 0.11 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr6_+_32936353 | 0.11 |
ENST00000374825.4 |
BRD2 |
bromodomain containing 2 |
| chr3_-_123339343 | 0.11 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr8_-_102181718 | 0.10 |
ENST00000565617.1 |
KB-1460A1.5 |
KB-1460A1.5 |
| chr20_-_13765526 | 0.10 |
ENST00000202816.1 |
ESF1 |
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr12_+_41831485 | 0.10 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr19_-_18433910 | 0.10 |
ENST00000594828.3 ENST00000593829.1 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr10_+_73156664 | 0.10 |
ENST00000398809.4 ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23 |
cadherin-related 23 |
| chr3_-_123339418 | 0.10 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chrX_+_47092314 | 0.10 |
ENST00000218348.3 |
USP11 |
ubiquitin specific peptidase 11 |
| chr1_+_117963209 | 0.10 |
ENST00000449370.2 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
| chr16_+_10837643 | 0.10 |
ENST00000574334.1 ENST00000283027.5 ENST00000433392.2 |
NUBP1 |
nucleotide binding protein 1 |
| chr1_-_36615051 | 0.10 |
ENST00000373163.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr5_-_179047881 | 0.10 |
ENST00000521173.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr11_-_72433346 | 0.10 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_+_120436733 | 0.09 |
ENST00000401466.1 ENST00000424086.1 ENST00000272521.6 |
TMEM177 |
transmembrane protein 177 |
| chr2_-_24270217 | 0.09 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chr5_+_167718604 | 0.09 |
ENST00000265293.4 |
WWC1 |
WW and C2 domain containing 1 |
| chr8_-_121457332 | 0.09 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chrM_+_12331 | 0.09 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr15_-_56757329 | 0.09 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr22_-_42086477 | 0.09 |
ENST00000402458.1 |
NHP2L1 |
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr5_+_159848807 | 0.09 |
ENST00000352433.5 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr8_+_110552337 | 0.09 |
ENST00000337573.5 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr5_+_125758865 | 0.09 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr12_-_71551652 | 0.09 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr1_+_202431859 | 0.08 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr21_-_36421401 | 0.08 |
ENST00000486278.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr2_-_27558270 | 0.08 |
ENST00000454704.1 |
GTF3C2 |
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr7_+_102715315 | 0.08 |
ENST00000428183.2 ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10 |
armadillo repeat containing 10 |
| chr5_-_11588907 | 0.08 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr5_-_58335281 | 0.08 |
ENST00000358923.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr12_+_14572070 | 0.08 |
ENST00000545769.1 ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr1_+_15272271 | 0.08 |
ENST00000400797.3 |
KAZN |
kazrin, periplakin interacting protein |
| chr15_-_42840961 | 0.08 |
ENST00000563454.1 ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57 |
leucine rich repeat containing 57 |
| chr6_-_11779014 | 0.07 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr5_+_125758813 | 0.07 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr17_-_2966901 | 0.07 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr7_-_22233442 | 0.07 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_110723335 | 0.07 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr6_-_150067632 | 0.07 |
ENST00000460354.2 ENST00000367404.4 ENST00000543637.1 |
NUP43 |
nucleoporin 43kDa |
| chr18_-_60985914 | 0.07 |
ENST00000589955.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr20_+_30697510 | 0.07 |
ENST00000217315.5 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr14_-_50154921 | 0.07 |
ENST00000553805.2 ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2 |
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr17_+_48823975 | 0.07 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr6_+_44355257 | 0.06 |
ENST00000371477.3 |
CDC5L |
cell division cycle 5-like |
| chr16_+_22501658 | 0.06 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr11_-_11374904 | 0.06 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr17_+_7591639 | 0.06 |
ENST00000396463.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr6_-_150067696 | 0.06 |
ENST00000340413.2 ENST00000367403.3 |
NUP43 |
nucleoporin 43kDa |
| chr20_+_30697298 | 0.06 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr2_+_162087577 | 0.06 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr1_+_150898812 | 0.06 |
ENST00000271640.5 ENST00000448029.1 ENST00000368962.2 ENST00000534805.1 ENST00000368969.4 ENST00000368963.1 ENST00000498193.1 |
SETDB1 |
SET domain, bifurcated 1 |
| chr8_-_133637624 | 0.06 |
ENST00000522789.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr19_+_47634039 | 0.06 |
ENST00000597808.1 ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
| chr6_+_45296391 | 0.06 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr8_-_22526597 | 0.06 |
ENST00000519513.1 ENST00000276416.6 ENST00000520292.1 ENST00000522268.1 |
BIN3 |
bridging integrator 3 |
| chr1_-_31538517 | 0.06 |
ENST00000440538.2 ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1 |
pumilio RNA-binding family member 1 |
| chr14_+_39703084 | 0.06 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr7_-_107443652 | 0.06 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr19_-_39881669 | 0.06 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr5_+_40909354 | 0.05 |
ENST00000313164.9 |
C7 |
complement component 7 |
| chr13_+_25875785 | 0.05 |
ENST00000381747.3 |
NUPL1 |
nucleoporin like 1 |
| chr18_-_60986613 | 0.05 |
ENST00000444484.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr9_+_123970052 | 0.05 |
ENST00000373823.3 |
GSN |
gelsolin |
| chr12_+_52056548 | 0.05 |
ENST00000545061.1 ENST00000355133.3 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
| chr2_+_162272605 | 0.05 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr13_+_25875662 | 0.05 |
ENST00000381736.3 ENST00000463407.1 ENST00000381718.3 |
NUPL1 |
nucleoporin like 1 |
| chr5_-_58652788 | 0.05 |
ENST00000405755.2 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr13_+_102104952 | 0.05 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr20_+_56964169 | 0.05 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr7_-_22234381 | 0.05 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr8_+_110552831 | 0.05 |
ENST00000530629.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr14_-_25078864 | 0.05 |
ENST00000216338.4 ENST00000557220.2 ENST00000382548.4 |
GZMH |
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr2_+_171034646 | 0.05 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr2_+_152266392 | 0.05 |
ENST00000444746.2 ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr2_-_217724767 | 0.05 |
ENST00000236979.2 |
TNP1 |
transition protein 1 (during histone to protamine replacement) |
| chr1_+_224301787 | 0.05 |
ENST00000366862.5 ENST00000424254.2 |
FBXO28 |
F-box protein 28 |
| chr6_+_20534672 | 0.05 |
ENST00000274695.4 ENST00000378624.4 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
| chr17_+_38171681 | 0.05 |
ENST00000225474.2 ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
| chr17_-_42143963 | 0.04 |
ENST00000585388.1 ENST00000293406.3 |
LSM12 |
LSM12 homolog (S. cerevisiae) |
| chr1_+_171217622 | 0.04 |
ENST00000433267.1 ENST00000367750.3 |
FMO1 |
flavin containing monooxygenase 1 |
| chr7_+_32535060 | 0.04 |
ENST00000318709.4 ENST00000409301.1 ENST00000404479.1 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
| chr1_-_160231451 | 0.04 |
ENST00000495887.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr18_+_9103957 | 0.04 |
ENST00000400033.1 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr1_+_211433275 | 0.04 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr17_+_7591747 | 0.04 |
ENST00000534050.1 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr4_+_113739244 | 0.04 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr9_-_86593238 | 0.04 |
ENST00000351839.3 |
HNRNPK |
heterogeneous nuclear ribonucleoprotein K |
| chrX_-_70473957 | 0.04 |
ENST00000373984.3 ENST00000314425.5 ENST00000373982.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr7_+_93535866 | 0.04 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr8_-_122653630 | 0.04 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr17_+_38171614 | 0.04 |
ENST00000583218.1 ENST00000394149.3 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
| chr8_-_121457608 | 0.04 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr5_-_11589131 | 0.04 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr5_-_98262240 | 0.04 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr9_+_71944241 | 0.04 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr2_-_40739501 | 0.04 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr4_+_144303093 | 0.04 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr6_+_45296048 | 0.04 |
ENST00000465038.2 ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr9_+_37753795 | 0.04 |
ENST00000377753.2 ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B |
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chrX_-_153363188 | 0.03 |
ENST00000303391.6 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr3_-_113897899 | 0.03 |
ENST00000383673.2 ENST00000295881.7 |
DRD3 |
dopamine receptor D3 |
| chr5_+_140552218 | 0.03 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr4_+_95679072 | 0.03 |
ENST00000515059.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr6_+_55039050 | 0.03 |
ENST00000370862.3 |
HCRTR2 |
hypocretin (orexin) receptor 2 |
| chr7_+_93535817 | 0.03 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr12_-_120315074 | 0.03 |
ENST00000261833.7 ENST00000392521.2 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
| chrX_-_130037198 | 0.03 |
ENST00000370935.1 ENST00000338144.3 ENST00000394363.1 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
| chr8_-_134072593 | 0.03 |
ENST00000427060.2 |
SLA |
Src-like-adaptor |
| chr1_+_50459990 | 0.03 |
ENST00000448346.1 |
AL645730.2 |
AL645730.2 |
| chr1_-_110155671 | 0.03 |
ENST00000351050.3 |
GNAT2 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr5_+_34757309 | 0.03 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr3_+_189507523 | 0.03 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr1_-_115259337 | 0.03 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr5_+_140602904 | 0.03 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr9_-_95166841 | 0.03 |
ENST00000262551.4 |
OGN |
osteoglycin |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.2 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.6 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |