Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NKX2-3
Z-value: 1.21
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NKX2-3 |
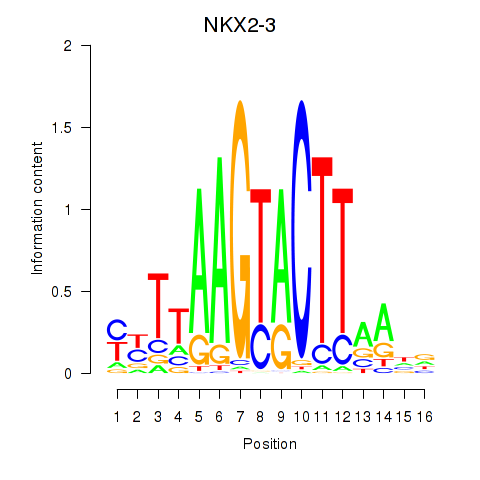
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150404904 | 7.68 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr11_+_10326612 | 4.45 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chrX_+_114795489 | 3.87 |
ENST00000355899.3 ENST00000537301.1 ENST00000289290.3 |
PLS3 |
plastin 3 |
| chr6_-_11382478 | 2.79 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_-_123512688 | 2.69 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr17_-_76870126 | 2.47 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr17_-_76870222 | 2.32 |
ENST00000585421.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr12_+_6309963 | 2.18 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr9_+_116263778 | 2.12 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr11_+_35211429 | 1.99 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr2_+_201170596 | 1.99 |
ENST00000439084.1 ENST00000409718.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr7_+_134430212 | 1.97 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr9_-_116840728 | 1.73 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr3_+_98482175 | 1.62 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr9_-_99382065 | 1.62 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr11_+_35211511 | 1.58 |
ENST00000524922.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr4_-_84035905 | 1.46 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr15_+_33022885 | 1.45 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr6_+_83073334 | 1.44 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr6_+_83072923 | 1.44 |
ENST00000535040.1 |
TPBG |
trophoblast glycoprotein |
| chr11_+_12399071 | 1.40 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr8_-_17555164 | 1.39 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr4_-_100242549 | 1.37 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr7_-_80548667 | 1.31 |
ENST00000265361.3 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr4_-_84035868 | 1.28 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr15_+_69452959 | 1.28 |
ENST00000261858.2 |
GLCE |
glucuronic acid epimerase |
| chr17_+_53344945 | 1.26 |
ENST00000575345.1 |
HLF |
hepatic leukemia factor |
| chr12_+_111051832 | 1.23 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr14_-_93214915 | 1.22 |
ENST00000553918.1 ENST00000555699.1 ENST00000553802.1 ENST00000554397.1 ENST00000554919.1 ENST00000554080.1 ENST00000553371.1 |
LGMN |
legumain |
| chr3_+_30647994 | 1.21 |
ENST00000295754.5 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr16_+_8736232 | 1.20 |
ENST00000562973.1 |
METTL22 |
methyltransferase like 22 |
| chr1_-_79472365 | 1.18 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chrX_-_134049262 | 1.16 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr12_+_111051902 | 1.16 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr3_-_122134882 | 1.15 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr10_+_135207598 | 1.13 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr16_+_69221028 | 1.13 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr3_+_30648066 | 1.12 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr9_+_36036430 | 1.07 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr2_-_38303218 | 1.03 |
ENST00000407341.1 ENST00000260630.3 |
CYP1B1 |
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr10_-_13523073 | 1.02 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr15_+_58702742 | 1.02 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr4_+_146403912 | 1.01 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr2_-_188419078 | 1.00 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr19_-_19051993 | 1.00 |
ENST00000594794.1 ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr10_+_135207623 | 0.96 |
ENST00000317502.6 ENST00000432508.3 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr17_+_36283971 | 0.94 |
ENST00000327454.6 ENST00000378174.5 |
TBC1D3F |
TBC1 domain family, member 3F |
| chr9_+_71820057 | 0.92 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr12_+_71833550 | 0.90 |
ENST00000266674.5 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr1_-_240775447 | 0.90 |
ENST00000318160.4 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
| chr9_+_71819927 | 0.90 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr13_-_46679185 | 0.89 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 0.88 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr8_-_93029865 | 0.82 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_36348610 | 0.82 |
ENST00000339023.4 ENST00000354664.4 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr17_-_34808047 | 0.82 |
ENST00000592614.1 ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G TBC1D3H |
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr2_-_188419200 | 0.81 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr16_-_18468926 | 0.80 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr17_+_66521936 | 0.79 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr20_-_4795747 | 0.78 |
ENST00000379376.2 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr2_-_36825281 | 0.76 |
ENST00000405912.3 ENST00000379245.4 |
FEZ2 |
fasciculation and elongation protein zeta 2 (zygin II) |
| chr3_-_47950745 | 0.76 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr4_-_104021009 | 0.76 |
ENST00000509245.1 ENST00000296424.4 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
| chr2_-_231989808 | 0.74 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr12_-_122296755 | 0.74 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr18_-_25616519 | 0.74 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr16_+_14802801 | 0.74 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr15_-_78369994 | 0.70 |
ENST00000300584.3 ENST00000409931.3 |
TBC1D2B |
TBC1 domain family, member 2B |
| chr8_-_49834299 | 0.69 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr20_+_57427765 | 0.69 |
ENST00000371100.4 |
GNAS |
GNAS complex locus |
| chr10_-_70092671 | 0.68 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr17_+_42264322 | 0.67 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr2_+_219110149 | 0.66 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr4_-_141677267 | 0.66 |
ENST00000442267.2 |
TBC1D9 |
TBC1 domain family, member 9 (with GRAM domain) |
| chr17_+_26662730 | 0.65 |
ENST00000226225.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_-_52002403 | 0.65 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr19_+_17622415 | 0.64 |
ENST00000252603.2 ENST00000600923.1 |
PGLS |
6-phosphogluconolactonase |
| chr3_-_49761337 | 0.63 |
ENST00000535833.1 ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3 GMPPB |
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr12_-_48398104 | 0.62 |
ENST00000337299.6 ENST00000380518.3 |
COL2A1 |
collagen, type II, alpha 1 |
| chr4_-_120550146 | 0.62 |
ENST00000354960.3 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
| chr8_-_49833978 | 0.61 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr7_-_2883928 | 0.61 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chr16_-_15474904 | 0.61 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr3_-_125802765 | 0.61 |
ENST00000514891.1 ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr1_+_54359854 | 0.60 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr7_-_87342564 | 0.58 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr20_-_33735070 | 0.57 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_-_123201337 | 0.56 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr4_-_57524061 | 0.54 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr13_+_50570019 | 0.54 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr4_+_113739244 | 0.53 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr13_-_50018241 | 0.53 |
ENST00000409308.1 |
CAB39L |
calcium binding protein 39-like |
| chr12_-_123187890 | 0.52 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr12_-_125398654 | 0.52 |
ENST00000541645.1 ENST00000540351.1 |
UBC |
ubiquitin C |
| chr16_+_56969284 | 0.52 |
ENST00000568358.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr17_+_26662597 | 0.51 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr14_-_60636561 | 0.51 |
ENST00000536410.2 ENST00000216500.5 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
| chr11_-_62389449 | 0.50 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr7_-_76829125 | 0.49 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr7_-_86849883 | 0.49 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr7_-_29186008 | 0.49 |
ENST00000396276.3 ENST00000265394.5 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr10_-_112678692 | 0.49 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr13_-_50018140 | 0.48 |
ENST00000410043.1 ENST00000347776.5 |
CAB39L |
calcium binding protein 39-like |
| chr17_-_26662464 | 0.47 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_44174330 | 0.47 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr6_-_137539651 | 0.46 |
ENST00000543628.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr13_-_44735393 | 0.44 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr17_-_26662440 | 0.42 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chrX_-_106362013 | 0.42 |
ENST00000372487.1 ENST00000372479.3 ENST00000203616.8 |
RBM41 |
RNA binding motif protein 41 |
| chr6_-_33239612 | 0.41 |
ENST00000482399.1 ENST00000445902.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr14_-_90798418 | 0.41 |
ENST00000354366.3 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
| chr6_-_33285505 | 0.40 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr14_-_75643296 | 0.40 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr9_+_118950325 | 0.39 |
ENST00000534838.1 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr19_-_39881669 | 0.38 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr14_-_95624227 | 0.37 |
ENST00000526495.1 |
DICER1 |
dicer 1, ribonuclease type III |
| chr20_+_33104199 | 0.35 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr12_-_120315074 | 0.34 |
ENST00000261833.7 ENST00000392521.2 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_+_202317815 | 0.33 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr18_-_51750948 | 0.33 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr4_-_16077741 | 0.32 |
ENST00000447510.2 ENST00000540805.1 ENST00000539194.1 |
PROM1 |
prominin 1 |
| chr20_-_23969416 | 0.31 |
ENST00000335694.4 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
| chr1_-_165668100 | 0.30 |
ENST00000354775.4 |
ALDH9A1 |
aldehyde dehydrogenase 9 family, member A1 |
| chr22_+_38004473 | 0.29 |
ENST00000414350.3 ENST00000343632.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr3_+_183967409 | 0.28 |
ENST00000324557.4 ENST00000402825.3 |
ECE2 |
endothelin converting enzyme 2 |
| chr1_-_2343951 | 0.28 |
ENST00000288774.3 ENST00000507596.1 ENST00000447513.2 |
PEX10 |
peroxisomal biogenesis factor 10 |
| chr2_+_173420697 | 0.28 |
ENST00000282077.3 ENST00000392571.2 ENST00000410055.1 |
PDK1 |
pyruvate dehydrogenase kinase, isozyme 1 |
| chr1_+_155278625 | 0.28 |
ENST00000368356.4 ENST00000356657.6 |
FDPS |
farnesyl diphosphate synthase |
| chr5_+_156693091 | 0.28 |
ENST00000318218.6 ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr19_+_13051206 | 0.27 |
ENST00000586760.1 |
CALR |
calreticulin |
| chr10_-_104262460 | 0.27 |
ENST00000446605.2 ENST00000369905.4 ENST00000545684.1 |
ACTR1A |
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr22_+_38453207 | 0.26 |
ENST00000404072.3 ENST00000424694.1 |
PICK1 |
protein interacting with PRKCA 1 |
| chr19_-_3772209 | 0.25 |
ENST00000555978.1 ENST00000555633.1 |
RAX2 |
retina and anterior neural fold homeobox 2 |
| chr18_+_61445007 | 0.25 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr3_+_149191723 | 0.25 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr5_-_142814241 | 0.25 |
ENST00000504572.1 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_+_71357744 | 0.25 |
ENST00000498451.2 |
MPHOSPH10 |
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr12_-_125398850 | 0.24 |
ENST00000535859.1 ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC |
ubiquitin C |
| chr11_-_14521379 | 0.24 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr1_+_155278539 | 0.24 |
ENST00000447866.1 |
FDPS |
farnesyl diphosphate synthase |
| chr12_+_14572070 | 0.23 |
ENST00000545769.1 ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr17_+_75401152 | 0.23 |
ENST00000585930.1 |
SEPT9 |
septin 9 |
| chr4_+_76439665 | 0.23 |
ENST00000508105.1 ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6 |
THAP domain containing 6 |
| chr11_+_124735282 | 0.22 |
ENST00000397801.1 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr3_-_114790179 | 0.21 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_-_69062764 | 0.21 |
ENST00000295571.5 |
EOGT |
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr5_+_49961727 | 0.20 |
ENST00000505697.2 ENST00000503750.2 ENST00000514342.2 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr3_-_150920979 | 0.19 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr1_+_158901329 | 0.19 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr2_+_64068844 | 0.19 |
ENST00000337130.5 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr7_-_150974494 | 0.18 |
ENST00000392811.2 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr12_+_49761273 | 0.18 |
ENST00000551540.1 ENST00000552918.1 ENST00000548777.1 ENST00000547865.1 ENST00000552171.1 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr12_+_49761224 | 0.17 |
ENST00000553127.1 ENST00000321898.6 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr19_+_15751689 | 0.16 |
ENST00000586182.2 ENST00000591058.1 ENST00000221307.8 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr6_+_46761118 | 0.16 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr2_+_161993412 | 0.16 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_+_147774275 | 0.15 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr6_+_33239787 | 0.15 |
ENST00000439602.2 ENST00000474973.1 |
RPS18 |
ribosomal protein S18 |
| chr19_-_42746714 | 0.15 |
ENST00000222330.3 |
GSK3A |
glycogen synthase kinase 3 alpha |
| chr14_+_23938891 | 0.15 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr6_-_33239712 | 0.15 |
ENST00000436044.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr5_-_115177247 | 0.15 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr4_+_88754113 | 0.14 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr18_-_64271363 | 0.14 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr9_-_115819039 | 0.14 |
ENST00000555206.1 |
ZFP37 |
ZFP37 zinc finger protein |
| chr17_+_29248698 | 0.14 |
ENST00000330889.3 |
ADAP2 |
ArfGAP with dual PH domains 2 |
| chr5_-_70320941 | 0.13 |
ENST00000523981.1 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr5_+_140552218 | 0.13 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr3_+_113775576 | 0.13 |
ENST00000485050.1 ENST00000281273.4 |
QTRTD1 |
queuine tRNA-ribosyltransferase domain containing 1 |
| chr11_-_111175739 | 0.13 |
ENST00000532918.1 |
COLCA1 |
colorectal cancer associated 1 |
| chr12_+_123464607 | 0.13 |
ENST00000543566.1 ENST00000315580.5 ENST00000542099.1 ENST00000392435.2 ENST00000413381.2 ENST00000426960.2 ENST00000453766.2 |
ARL6IP4 |
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr19_-_1401486 | 0.12 |
ENST00000252288.2 ENST00000447102.3 |
GAMT |
guanidinoacetate N-methyltransferase |
| chr10_-_32345305 | 0.12 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chr6_+_168399772 | 0.11 |
ENST00000443060.2 |
KIF25 |
kinesin family member 25 |
| chr10_-_104262426 | 0.11 |
ENST00000487599.1 |
ACTR1A |
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr2_+_234590556 | 0.11 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr3_+_186358200 | 0.10 |
ENST00000382136.3 |
FETUB |
fetuin B |
| chr5_+_149569520 | 0.10 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr8_+_134125727 | 0.10 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr18_-_33702078 | 0.10 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr10_+_17794251 | 0.10 |
ENST00000377495.1 ENST00000338221.5 |
TMEM236 |
transmembrane protein 236 |
| chr10_+_18041218 | 0.10 |
ENST00000480516.1 ENST00000457860.1 |
TMEM236 |
TMEM236 |
| chr19_+_42746927 | 0.09 |
ENST00000378108.1 |
AC006486.1 |
AC006486.1 |
| chr12_+_56546223 | 0.09 |
ENST00000550443.1 ENST00000207437.5 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr18_+_32173276 | 0.08 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr17_-_8055747 | 0.08 |
ENST00000317276.4 ENST00000581703.1 |
PER1 |
period circadian clock 1 |
| chr16_-_11363178 | 0.08 |
ENST00000312693.3 |
TNP2 |
transition protein 2 (during histone to protamine replacement) |
| chr2_+_89923550 | 0.08 |
ENST00000509129.1 |
IGKV1D-37 |
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr11_+_11863500 | 0.08 |
ENST00000527733.1 ENST00000539466.1 |
USP47 |
ubiquitin specific peptidase 47 |
| chr11_+_28131821 | 0.08 |
ENST00000379199.2 ENST00000303459.6 |
METTL15 |
methyltransferase like 15 |
| chr19_+_45409011 | 0.07 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr20_+_55043647 | 0.07 |
ENST00000023939.4 ENST00000395881.3 ENST00000357348.5 ENST00000449062.1 ENST00000435342.2 |
RTFDC1 |
replication termination factor 2 domain containing 1 |
| chr1_-_35325400 | 0.07 |
ENST00000521580.2 |
SMIM12 |
small integral membrane protein 12 |
| chr8_-_20040601 | 0.07 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr6_+_32006042 | 0.07 |
ENST00000418967.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_-_75695366 | 0.07 |
ENST00000512743.1 |
BTC |
betacellulin |
| chr20_+_60962143 | 0.07 |
ENST00000343986.4 |
RPS21 |
ribosomal protein S21 |
| chr3_-_167191814 | 0.07 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr6_-_111927062 | 0.06 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr2_-_89597542 | 0.06 |
ENST00000465170.1 |
IGKV1-37 |
immunoglobulin kappa variable 1-37 (non-functional) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 0.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.7 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 2.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 2.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 4.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 4.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.7 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 16.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 4.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 5.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 4.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 4.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.8 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.0 | 4.8 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.9 | 4.5 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.8 | 2.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.6 | 2.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 1.4 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 3.6 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 1.8 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 1.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.4 | 1.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.3 | 2.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 0.9 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 1.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 0.6 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 2.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 2.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 0.6 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 0.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 1.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.7 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.5 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.1 | 1.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 1.7 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 1.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 1.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.5 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.9 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 1.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.5 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 1.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.6 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 2.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.3 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.3 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 1.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 3.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 2.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.6 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 2.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.3 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.5 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0046498 | creatine metabolic process(GO:0006600) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 2.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 2.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 1.6 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.3 | 0.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.9 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 3.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.7 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.2 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 1.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |