Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NKX2-4
Z-value: 0.64
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
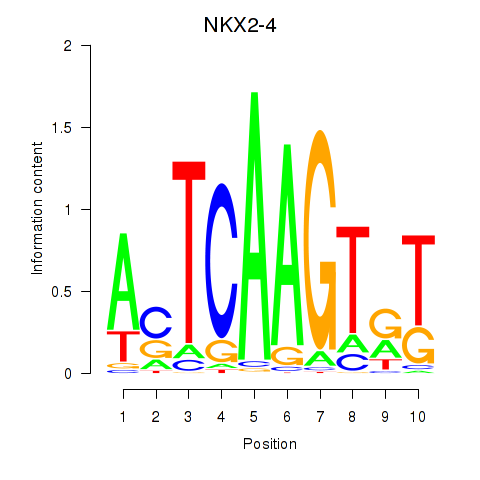
NKX2-4
|
ENSG00000125816.4 | NKX2-4 |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_15918618 | 1.94 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chrX_-_154563889 | 1.90 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr3_-_107777208 | 1.35 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr11_+_60223225 | 1.34 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_60223312 | 1.29 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chrX_+_49294472 | 1.08 |
ENST00000361446.5 |
GAGE12B |
G antigen 12B |
| chr10_-_121296045 | 1.07 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr15_-_55541227 | 0.99 |
ENST00000566877.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chrX_+_49178536 | 0.82 |
ENST00000442437.2 |
GAGE12J |
G antigen 12J |
| chr17_-_29641104 | 0.80 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr16_+_12059091 | 0.79 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr6_+_12958137 | 0.79 |
ENST00000457702.2 ENST00000379345.2 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr12_+_48516357 | 0.77 |
ENST00000549022.1 ENST00000547587.1 ENST00000312352.7 |
PFKM |
phosphofructokinase, muscle |
| chr6_+_26158343 | 0.75 |
ENST00000377777.4 ENST00000289316.2 |
HIST1H2BD |
histone cluster 1, H2bd |
| chr1_+_161632937 | 0.75 |
ENST00000236937.9 ENST00000367961.4 ENST00000358671.5 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chrX_+_78200913 | 0.74 |
ENST00000171757.2 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chrX_+_78200829 | 0.74 |
ENST00000544091.1 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr15_-_20193370 | 0.70 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr14_+_75988851 | 0.67 |
ENST00000555504.1 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr20_-_43280325 | 0.66 |
ENST00000537820.1 |
ADA |
adenosine deaminase |
| chr16_+_28943260 | 0.62 |
ENST00000538922.1 ENST00000324662.3 ENST00000567541.1 |
CD19 |
CD19 molecule |
| chr17_-_29641084 | 0.61 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr11_-_5255861 | 0.61 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr5_-_176836577 | 0.59 |
ENST00000253496.3 |
F12 |
coagulation factor XII (Hageman factor) |
| chr16_+_1730338 | 0.58 |
ENST00000566691.1 ENST00000382710.4 |
HN1L |
hematological and neurological expressed 1-like |
| chr15_+_75315896 | 0.57 |
ENST00000342932.3 ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr5_-_34919094 | 0.57 |
ENST00000341754.4 |
RAD1 |
RAD1 homolog (S. pombe) |
| chr7_+_116139424 | 0.55 |
ENST00000222693.4 |
CAV2 |
caveolin 2 |
| chr12_+_10658201 | 0.54 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_+_32716857 | 0.53 |
ENST00000482949.1 ENST00000495610.2 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr19_-_55669093 | 0.52 |
ENST00000344887.5 |
TNNI3 |
troponin I type 3 (cardiac) |
| chrX_-_124097620 | 0.52 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr1_+_15986364 | 0.51 |
ENST00000345034.1 |
RSC1A1 |
regulatory solute carrier protein, family 1, member 1 |
| chr10_+_35484053 | 0.51 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr9_+_93589734 | 0.48 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr11_+_118958689 | 0.45 |
ENST00000535253.1 ENST00000392841.1 |
HMBS |
hydroxymethylbilane synthase |
| chrX_-_110655306 | 0.45 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr12_-_53601055 | 0.45 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr1_-_197036364 | 0.44 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr11_-_19263145 | 0.44 |
ENST00000532666.1 ENST00000527884.1 |
E2F8 |
E2F transcription factor 8 |
| chrX_+_48542168 | 0.43 |
ENST00000376701.4 |
WAS |
Wiskott-Aldrich syndrome |
| chr13_+_111767650 | 0.43 |
ENST00000449979.1 ENST00000370623.3 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr22_-_26961328 | 0.42 |
ENST00000398110.2 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
| chr6_+_5261225 | 0.42 |
ENST00000324331.6 |
FARS2 |
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr11_-_19223523 | 0.41 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr14_-_23451467 | 0.41 |
ENST00000555074.1 ENST00000361265.4 |
RP11-298I3.5 AJUBA |
RP11-298I3.5 ajuba LIM protein |
| chr7_-_7782204 | 0.41 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chr12_-_53601000 | 0.41 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr19_-_6670128 | 0.40 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr6_+_83073334 | 0.40 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr11_-_62439012 | 0.39 |
ENST00000532208.1 ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48 |
chromosome 11 open reading frame 48 |
| chr19_-_20748541 | 0.38 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr5_-_88119580 | 0.38 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_-_225434538 | 0.37 |
ENST00000409096.1 |
CUL3 |
cullin 3 |
| chr19_-_11688500 | 0.37 |
ENST00000433365.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr2_-_175462456 | 0.37 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr6_+_83072923 | 0.36 |
ENST00000535040.1 |
TPBG |
trophoblast glycoprotein |
| chr8_-_28747424 | 0.36 |
ENST00000523436.1 ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9 |
integrator complex subunit 9 |
| chr19_+_41117770 | 0.36 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr9_+_134065506 | 0.35 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr16_+_31539197 | 0.35 |
ENST00000564707.1 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr12_-_102591604 | 0.34 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr19_+_44100544 | 0.34 |
ENST00000391965.2 ENST00000525771.1 |
ZNF576 |
zinc finger protein 576 |
| chr6_+_35265586 | 0.33 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr1_+_161551101 | 0.32 |
ENST00000367962.4 ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr14_+_74353320 | 0.31 |
ENST00000540593.1 ENST00000555730.1 |
ZNF410 |
zinc finger protein 410 |
| chr16_+_31539183 | 0.31 |
ENST00000302312.4 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr5_-_11588907 | 0.31 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr18_+_21032781 | 0.31 |
ENST00000339486.3 |
RIOK3 |
RIO kinase 3 |
| chr2_-_175462934 | 0.31 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr3_-_49066811 | 0.30 |
ENST00000442157.1 ENST00000326739.4 |
IMPDH2 |
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_-_85780853 | 0.30 |
ENST00000531930.1 ENST00000528398.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_44100632 | 0.30 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
| chr9_-_32573130 | 0.30 |
ENST00000350021.2 ENST00000379847.3 |
NDUFB6 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chrX_-_110655391 | 0.29 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr1_-_153643442 | 0.29 |
ENST00000368681.1 ENST00000361891.4 |
ILF2 |
interleukin enhancer binding factor 2 |
| chr18_-_28622699 | 0.29 |
ENST00000360428.4 |
DSC3 |
desmocollin 3 |
| chr10_+_105005644 | 0.28 |
ENST00000441178.2 |
RP11-332O19.5 |
ribulose-5-phosphate-3-epimerase-like 1 |
| chr7_+_120629653 | 0.28 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr20_-_4804244 | 0.28 |
ENST00000379400.3 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr12_+_123011776 | 0.27 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr12_-_56843161 | 0.27 |
ENST00000554616.1 ENST00000553532.1 ENST00000229201.4 |
TIMELESS |
timeless circadian clock |
| chr12_+_58176525 | 0.27 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr18_-_33702078 | 0.27 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr3_+_118905564 | 0.27 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chrX_-_133792480 | 0.26 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr5_+_150040403 | 0.26 |
ENST00000517768.1 ENST00000297130.4 |
MYOZ3 |
myozenin 3 |
| chr12_-_104532062 | 0.26 |
ENST00000240055.3 |
NFYB |
nuclear transcription factor Y, beta |
| chr10_+_22605374 | 0.25 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr10_-_28571015 | 0.25 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_+_123237321 | 0.25 |
ENST00000280557.6 ENST00000455982.2 |
DENR |
density-regulated protein |
| chr4_-_87028478 | 0.25 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr12_+_56435637 | 0.25 |
ENST00000356464.5 ENST00000552361.1 |
RPS26 |
ribosomal protein S26 |
| chr21_+_40823753 | 0.25 |
ENST00000333634.4 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr10_-_61900762 | 0.25 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_+_646960 | 0.25 |
ENST00000488061.1 ENST00000429163.2 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr1_+_161136180 | 0.25 |
ENST00000352210.5 ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX |
protoporphyrinogen oxidase |
| chr14_-_71067360 | 0.24 |
ENST00000554963.1 ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6 |
mediator complex subunit 6 |
| chr3_+_4535155 | 0.24 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr18_-_28622774 | 0.24 |
ENST00000434452.1 |
DSC3 |
desmocollin 3 |
| chr10_-_69455873 | 0.24 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr3_+_15045419 | 0.24 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr9_+_131445928 | 0.23 |
ENST00000372692.4 |
SET |
SET nuclear oncogene |
| chr3_+_189349162 | 0.23 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr5_+_159436120 | 0.23 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr17_+_60501228 | 0.23 |
ENST00000311506.5 |
METTL2A |
methyltransferase like 2A |
| chr1_+_144989309 | 0.23 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr4_+_39699664 | 0.22 |
ENST00000261427.5 ENST00000510934.1 ENST00000295963.6 |
UBE2K |
ubiquitin-conjugating enzyme E2K |
| chr6_-_149969871 | 0.22 |
ENST00000335643.8 ENST00000444282.1 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr12_+_50451331 | 0.21 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chrX_+_148622138 | 0.21 |
ENST00000450602.2 ENST00000441248.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr7_+_117120017 | 0.21 |
ENST00000003084.6 ENST00000454343.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr10_+_22605304 | 0.21 |
ENST00000475460.2 ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1 COMMD3 |
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr6_-_29343068 | 0.21 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr12_-_71182695 | 0.21 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr14_+_88471468 | 0.21 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr17_-_40264692 | 0.20 |
ENST00000591220.1 ENST00000251642.3 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr1_+_95583479 | 0.20 |
ENST00000455656.1 ENST00000604534.1 |
TMEM56 RP11-57H12.6 |
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr5_+_43602750 | 0.20 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr3_+_190105909 | 0.20 |
ENST00000456423.1 |
CLDN16 |
claudin 16 |
| chr7_+_75932863 | 0.20 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr1_-_89458287 | 0.20 |
ENST00000370485.2 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
| chr9_+_140083099 | 0.20 |
ENST00000322310.5 |
SSNA1 |
Sjogren syndrome nuclear autoantigen 1 |
| chr12_+_96252706 | 0.19 |
ENST00000266735.5 ENST00000553192.1 ENST00000552085.1 |
SNRPF |
small nuclear ribonucleoprotein polypeptide F |
| chr2_-_176046391 | 0.19 |
ENST00000392541.3 ENST00000409194.1 |
ATP5G3 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr16_+_81272287 | 0.19 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr6_-_149969829 | 0.19 |
ENST00000367411.2 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr18_+_29769978 | 0.19 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr14_+_39703112 | 0.18 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr17_-_62493131 | 0.18 |
ENST00000539111.2 |
POLG2 |
polymerase (DNA directed), gamma 2, accessory subunit |
| chr1_-_85040090 | 0.18 |
ENST00000370630.5 |
CTBS |
chitobiase, di-N-acetyl- |
| chr2_-_166651191 | 0.18 |
ENST00000392701.3 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr6_+_27925019 | 0.17 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr11_+_19138670 | 0.17 |
ENST00000446113.2 ENST00000399351.3 |
ZDHHC13 |
zinc finger, DHHC-type containing 13 |
| chr2_-_219925189 | 0.17 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr2_-_37899323 | 0.17 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_+_140235469 | 0.17 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr19_+_13135386 | 0.17 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_-_86849883 | 0.16 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr2_-_37544209 | 0.16 |
ENST00000234179.2 |
PRKD3 |
protein kinase D3 |
| chr12_+_54422142 | 0.16 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr1_-_156721502 | 0.16 |
ENST00000357325.5 |
HDGF |
hepatoma-derived growth factor |
| chr1_+_147374915 | 0.16 |
ENST00000240986.4 |
GJA8 |
gap junction protein, alpha 8, 50kDa |
| chr7_+_107384142 | 0.16 |
ENST00000440859.3 |
CBLL1 |
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr6_+_152011628 | 0.16 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr11_+_62439126 | 0.16 |
ENST00000377953.3 |
C11orf83 |
chromosome 11 open reading frame 83 |
| chr19_-_41942344 | 0.16 |
ENST00000594660.1 |
ATP5SL |
ATP5S-like |
| chr1_+_46769303 | 0.16 |
ENST00000311672.5 |
UQCRH |
ubiquinol-cytochrome c reductase hinge protein |
| chr1_+_117297007 | 0.15 |
ENST00000369478.3 ENST00000369477.1 |
CD2 |
CD2 molecule |
| chr7_+_117120106 | 0.15 |
ENST00000426809.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr4_-_175443943 | 0.15 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr16_-_3450963 | 0.15 |
ENST00000573327.1 ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32 |
zinc finger and SCAN domain containing 32 |
| chr1_-_155271213 | 0.15 |
ENST00000342741.4 |
PKLR |
pyruvate kinase, liver and RBC |
| chr11_-_119993979 | 0.15 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr13_-_36944307 | 0.15 |
ENST00000355182.4 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr9_-_95244781 | 0.15 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr6_-_41703952 | 0.15 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr7_-_55606346 | 0.15 |
ENST00000545390.1 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr15_+_40987327 | 0.15 |
ENST00000423169.2 ENST00000267868.3 ENST00000557850.1 ENST00000532743.1 ENST00000382643.3 |
RAD51 |
RAD51 recombinase |
| chr11_-_85779971 | 0.15 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_119445306 | 0.14 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr2_+_166326157 | 0.14 |
ENST00000421875.1 ENST00000314499.7 ENST00000409664.1 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr7_+_65552756 | 0.14 |
ENST00000450043.1 |
AC068533.7 |
AC068533.7 |
| chr12_-_322504 | 0.14 |
ENST00000424061.2 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr19_+_36235964 | 0.14 |
ENST00000587708.2 |
PSENEN |
presenilin enhancer gamma secretase subunit |
| chr7_-_86849025 | 0.14 |
ENST00000257637.3 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr7_-_92463210 | 0.14 |
ENST00000265734.4 |
CDK6 |
cyclin-dependent kinase 6 |
| chr10_+_81272287 | 0.13 |
ENST00000520547.2 |
EIF5AL1 |
eukaryotic translation initiation factor 5A-like 1 |
| chr19_+_37998031 | 0.13 |
ENST00000586138.1 ENST00000588578.1 ENST00000587986.1 |
ZNF793 |
zinc finger protein 793 |
| chr4_+_109541772 | 0.13 |
ENST00000506397.1 ENST00000394668.2 |
RPL34 |
ribosomal protein L34 |
| chr10_+_21823079 | 0.13 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr4_+_96012614 | 0.13 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr4_-_69434245 | 0.13 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr4_-_100242549 | 0.13 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr10_-_11574274 | 0.13 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr1_-_110155671 | 0.13 |
ENST00000351050.3 |
GNAT2 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr4_-_168155169 | 0.13 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_27863706 | 0.12 |
ENST00000081029.3 ENST00000538315.1 ENST00000542791.1 |
MRPS35 |
mitochondrial ribosomal protein S35 |
| chrX_+_49091920 | 0.12 |
ENST00000376227.3 |
CCDC22 |
coiled-coil domain containing 22 |
| chr7_-_14028488 | 0.12 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr2_+_89998789 | 0.12 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr14_+_65381079 | 0.12 |
ENST00000549115.1 ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1 FNTB CHURC1-FNTB |
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr1_+_70671363 | 0.12 |
ENST00000370951.1 ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11 |
serine/arginine-rich splicing factor 11 |
| chr4_+_75480629 | 0.12 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr4_-_68411275 | 0.12 |
ENST00000273853.6 |
CENPC |
centromere protein C |
| chr12_+_12878829 | 0.12 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr2_-_179315786 | 0.12 |
ENST00000457633.1 ENST00000438687.3 ENST00000325748.4 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr15_-_88799661 | 0.12 |
ENST00000360948.2 ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr11_-_506316 | 0.12 |
ENST00000532055.1 ENST00000531540.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr1_-_203198790 | 0.12 |
ENST00000367229.1 ENST00000255427.3 ENST00000535569.1 |
CHIT1 |
chitinase 1 (chitotriosidase) |
| chr1_-_70671216 | 0.11 |
ENST00000370952.3 |
LRRC40 |
leucine rich repeat containing 40 |
| chr6_-_31088214 | 0.11 |
ENST00000376288.2 |
CDSN |
corneodesmosin |
| chr8_-_74206673 | 0.11 |
ENST00000396465.1 |
RPL7 |
ribosomal protein L7 |
| chr12_+_21590549 | 0.11 |
ENST00000545178.1 ENST00000240651.9 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr19_+_45504688 | 0.11 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_-_111383064 | 0.11 |
ENST00000525791.1 ENST00000456861.2 ENST00000356018.2 |
BTG4 |
B-cell translocation gene 4 |
| chr5_+_140227048 | 0.11 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr11_-_106889250 | 0.11 |
ENST00000526355.2 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
| chr4_+_111397216 | 0.10 |
ENST00000265162.5 |
ENPEP |
glutamyl aminopeptidase (aminopeptidase A) |
| chr17_-_67138015 | 0.10 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0022829 | gap junction channel activity(GO:0005243) wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0043318 | follicular B cell differentiation(GO:0002316) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.3 | 0.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 0.7 | GO:0046102 | hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) |
| 0.2 | 0.9 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 0.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 1.0 | GO:0043316 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.9 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.4 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.5 | GO:0042223 | response to molecule of fungal origin(GO:0002238) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.4 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.4 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 1.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.4 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.5 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 2.0 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.7 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 2.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:2000314 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.5 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.1 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.7 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0045636 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 1.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.7 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |