Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
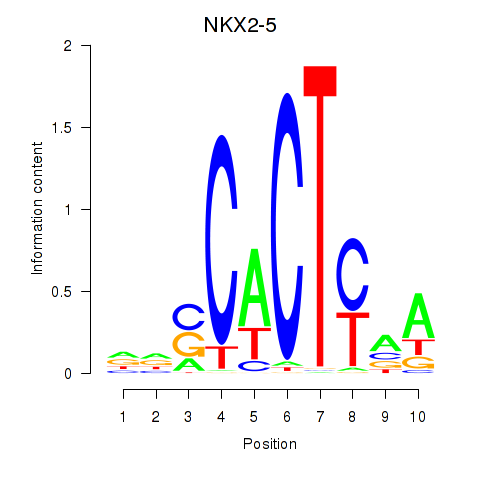
Results for NKX2-5
Z-value: 0.52
Transcription factors associated with NKX2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-5
|
ENSG00000183072.9 | NKX2-5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-5 | hg19_v2_chr5_-_172662303_172662360 | -0.41 | 1.2e-01 | Click! |
Activity profile of NKX2-5 motif
Sorted Z-values of NKX2-5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_149792295 | 0.96 |
ENST00000518797.1 ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr14_-_106406090 | 0.95 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chr17_-_29641104 | 0.85 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr17_-_29641084 | 0.69 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr14_-_106963409 | 0.66 |
ENST00000390621.2 |
IGHV1-45 |
immunoglobulin heavy variable 1-45 |
| chr12_+_113354341 | 0.63 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_+_99775366 | 0.60 |
ENST00000394018.2 ENST00000416412.1 |
STAG3 |
stromal antigen 3 |
| chr7_+_99775520 | 0.58 |
ENST00000317296.5 ENST00000422690.1 ENST00000439782.1 |
STAG3 |
stromal antigen 3 |
| chr13_-_41240717 | 0.55 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr5_+_75699040 | 0.53 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr15_+_81589254 | 0.51 |
ENST00000394652.2 |
IL16 |
interleukin 16 |
| chr2_-_175499294 | 0.50 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr8_-_120651020 | 0.46 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr17_+_26646175 | 0.44 |
ENST00000583381.1 ENST00000582113.1 ENST00000582384.1 |
TMEM97 |
transmembrane protein 97 |
| chr1_+_186798073 | 0.41 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr17_+_26646121 | 0.39 |
ENST00000226230.6 |
TMEM97 |
transmembrane protein 97 |
| chr7_-_37488834 | 0.39 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr3_-_15374659 | 0.34 |
ENST00000426925.1 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr17_+_48624450 | 0.33 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr5_-_171615315 | 0.32 |
ENST00000176763.5 |
STK10 |
serine/threonine kinase 10 |
| chr5_+_122110691 | 0.30 |
ENST00000379516.2 ENST00000505934.1 ENST00000514949.1 |
SNX2 |
sorting nexin 2 |
| chr22_+_40322595 | 0.29 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr22_+_40322623 | 0.29 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr11_+_108093839 | 0.28 |
ENST00000452508.2 |
ATM |
ataxia telangiectasia mutated |
| chr3_-_111314230 | 0.28 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr9_-_34048873 | 0.27 |
ENST00000449054.1 ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2 |
ubiquitin associated protein 2 |
| chr12_+_93772402 | 0.27 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr1_-_55352834 | 0.25 |
ENST00000371269.3 |
DHCR24 |
24-dehydrocholesterol reductase |
| chrX_-_154299501 | 0.23 |
ENST00000369476.3 ENST00000369484.3 |
MTCP1 CMC4 |
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr4_+_2470664 | 0.23 |
ENST00000314289.8 ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4 |
ring finger protein 4 |
| chr18_+_42260861 | 0.22 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr1_+_27153173 | 0.22 |
ENST00000374142.4 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
| chr3_-_56717246 | 0.21 |
ENST00000355628.5 |
FAM208A |
family with sequence similarity 208, member A |
| chr3_-_11685345 | 0.21 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr5_-_76788317 | 0.20 |
ENST00000296679.4 |
WDR41 |
WD repeat domain 41 |
| chr15_+_42841008 | 0.20 |
ENST00000260372.3 ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2 |
HAUS augmin-like complex, subunit 2 |
| chr1_-_22109682 | 0.19 |
ENST00000400301.1 ENST00000532737.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr16_+_77233294 | 0.18 |
ENST00000378644.4 |
SYCE1L |
synaptonemal complex central element protein 1-like |
| chr11_+_64879317 | 0.18 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr20_+_57226284 | 0.17 |
ENST00000458280.1 ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16 |
syntaxin 16 |
| chr10_+_76586348 | 0.17 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr12_+_56360550 | 0.17 |
ENST00000266970.4 |
CDK2 |
cyclin-dependent kinase 2 |
| chr9_-_117692697 | 0.17 |
ENST00000223795.2 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
| chr16_+_33006369 | 0.16 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr20_+_57226841 | 0.16 |
ENST00000358029.4 ENST00000361830.3 |
STX16 |
syntaxin 16 |
| chr19_+_5681153 | 0.15 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr11_+_18417813 | 0.15 |
ENST00000540430.1 ENST00000379412.5 |
LDHA |
lactate dehydrogenase A |
| chr6_-_41715128 | 0.14 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr17_+_53342311 | 0.14 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr10_+_73156664 | 0.13 |
ENST00000398809.4 ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23 |
cadherin-related 23 |
| chr7_-_87342564 | 0.13 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr17_-_29624343 | 0.13 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr3_+_42201653 | 0.12 |
ENST00000341421.3 ENST00000396175.1 |
TRAK1 |
trafficking protein, kinesin binding 1 |
| chr12_+_93772326 | 0.12 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chrX_-_14047996 | 0.12 |
ENST00000380523.4 ENST00000398355.3 |
GEMIN8 |
gem (nuclear organelle) associated protein 8 |
| chrX_+_115567767 | 0.12 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr8_+_76452097 | 0.12 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr8_-_95220775 | 0.11 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr7_-_81399355 | 0.11 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr19_+_5681011 | 0.11 |
ENST00000581893.1 ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L |
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr2_+_201997492 | 0.11 |
ENST00000494258.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr15_-_74726283 | 0.11 |
ENST00000543145.2 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr19_+_1026566 | 0.11 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr11_+_43702236 | 0.11 |
ENST00000531185.1 ENST00000278353.4 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr22_-_38577782 | 0.10 |
ENST00000430886.1 ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr7_-_81399438 | 0.10 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399329 | 0.10 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_+_133753533 | 0.10 |
ENST00000422256.2 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr17_+_75447326 | 0.10 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr12_-_371994 | 0.10 |
ENST00000343164.4 ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13 |
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr22_-_38577731 | 0.09 |
ENST00000335539.3 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr11_-_19223523 | 0.09 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr15_-_42840961 | 0.09 |
ENST00000563454.1 ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57 |
leucine rich repeat containing 57 |
| chr17_+_29421987 | 0.09 |
ENST00000431387.4 |
NF1 |
neurofibromin 1 |
| chr7_+_129007964 | 0.09 |
ENST00000460109.1 ENST00000474594.1 ENST00000446212.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr17_-_3500392 | 0.09 |
ENST00000571088.1 ENST00000174621.6 |
TRPV1 |
transient receptor potential cation channel, subfamily V, member 1 |
| chr7_-_81399411 | 0.09 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_101841588 | 0.08 |
ENST00000370418.3 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr15_+_42696992 | 0.08 |
ENST00000561817.1 |
CAPN3 |
calpain 3, (p94) |
| chr4_+_81118647 | 0.08 |
ENST00000415738.2 |
PRDM8 |
PR domain containing 8 |
| chr8_+_48873453 | 0.08 |
ENST00000523944.1 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr6_-_112115074 | 0.08 |
ENST00000368667.2 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr15_-_81282133 | 0.08 |
ENST00000261758.4 |
MESDC2 |
mesoderm development candidate 2 |
| chr17_+_40811283 | 0.08 |
ENST00000251412.7 |
TUBG2 |
tubulin, gamma 2 |
| chr1_-_11907829 | 0.08 |
ENST00000376480.3 |
NPPA |
natriuretic peptide A |
| chr15_+_42697018 | 0.08 |
ENST00000397204.4 |
CAPN3 |
calpain 3, (p94) |
| chr2_-_220041617 | 0.07 |
ENST00000451647.1 ENST00000360507.5 |
CNPPD1 |
cyclin Pas1/PHO80 domain containing 1 |
| chr18_+_61575200 | 0.07 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr19_-_5680499 | 0.07 |
ENST00000587589.1 |
C19orf70 |
chromosome 19 open reading frame 70 |
| chr8_+_48873479 | 0.07 |
ENST00000262105.2 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr8_+_132916318 | 0.07 |
ENST00000254624.5 ENST00000522709.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr13_+_25670268 | 0.07 |
ENST00000281589.3 |
PABPC3 |
poly(A) binding protein, cytoplasmic 3 |
| chr19_+_16999654 | 0.07 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr16_+_8814563 | 0.07 |
ENST00000425191.2 ENST00000569156.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr4_+_113970772 | 0.07 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr3_-_187388173 | 0.07 |
ENST00000287641.3 |
SST |
somatostatin |
| chr7_+_142457315 | 0.07 |
ENST00000486171.1 ENST00000311737.7 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr5_+_140345820 | 0.07 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr12_-_49259643 | 0.07 |
ENST00000309739.5 |
RND1 |
Rho family GTPase 1 |
| chr9_+_33265011 | 0.06 |
ENST00000419016.2 |
CHMP5 |
charged multivesicular body protein 5 |
| chr9_+_118950325 | 0.06 |
ENST00000534838.1 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr21_+_44313375 | 0.06 |
ENST00000354250.2 ENST00000340344.4 |
NDUFV3 |
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr1_+_211433275 | 0.06 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr15_+_42696954 | 0.05 |
ENST00000337571.4 ENST00000569136.1 |
CAPN3 |
calpain 3, (p94) |
| chr3_+_23847394 | 0.05 |
ENST00000306627.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr17_-_7493390 | 0.05 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr3_+_45984947 | 0.05 |
ENST00000304552.4 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
| chr20_-_44485835 | 0.05 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr8_-_10588010 | 0.05 |
ENST00000304501.1 |
SOX7 |
SRY (sex determining region Y)-box 7 |
| chr7_-_128415844 | 0.05 |
ENST00000249389.2 |
OPN1SW |
opsin 1 (cone pigments), short-wave-sensitive |
| chr17_+_38333263 | 0.04 |
ENST00000456989.2 ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr10_+_75504105 | 0.04 |
ENST00000535742.1 ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C |
SEC24 family member C |
| chr1_-_113498943 | 0.04 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr1_-_115259337 | 0.03 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr17_+_58755184 | 0.03 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr12_+_14518598 | 0.03 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr4_-_74853897 | 0.03 |
ENST00000296028.3 |
PPBP |
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr6_-_94129244 | 0.03 |
ENST00000369303.4 ENST00000369297.1 |
EPHA7 |
EPH receptor A7 |
| chr22_-_50524298 | 0.03 |
ENST00000311597.5 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr11_-_108464321 | 0.03 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr11_+_120973375 | 0.03 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr22_-_50523760 | 0.03 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr2_-_96874553 | 0.03 |
ENST00000337288.5 ENST00000443962.1 |
STARD7 |
StAR-related lipid transfer (START) domain containing 7 |
| chr2_+_152214098 | 0.03 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_201936302 | 0.02 |
ENST00000453765.1 ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B |
family with sequence similarity 126, member B |
| chr1_+_32671236 | 0.02 |
ENST00000537469.1 ENST00000291358.6 |
IQCC |
IQ motif containing C |
| chr2_+_219646462 | 0.02 |
ENST00000258415.4 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr9_+_33264861 | 0.02 |
ENST00000223500.8 |
CHMP5 |
charged multivesicular body protein 5 |
| chr6_+_31916733 | 0.02 |
ENST00000483004.1 |
CFB |
complement factor B |
| chr13_-_95364389 | 0.02 |
ENST00000376945.2 |
SOX21 |
SRY (sex determining region Y)-box 21 |
| chr3_+_23847432 | 0.02 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr10_-_81320151 | 0.02 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chr11_+_76813886 | 0.02 |
ENST00000529803.1 |
OMP |
olfactory marker protein |
| chr11_-_108464465 | 0.02 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr2_-_27545431 | 0.02 |
ENST00000233545.2 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr2_+_136289030 | 0.02 |
ENST00000409478.1 ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1 |
R3H domain containing 1 |
| chr17_+_27055798 | 0.02 |
ENST00000268766.6 |
NEK8 |
NIMA-related kinase 8 |
| chr9_+_116263639 | 0.02 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr3_+_108321623 | 0.02 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr12_+_4918342 | 0.01 |
ENST00000280684.3 ENST00000433855.1 |
KCNA6 |
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr14_+_22963806 | 0.01 |
ENST00000390493.1 |
TRAJ44 |
T cell receptor alpha joining 44 |
| chr5_+_74062806 | 0.01 |
ENST00000296802.5 |
NSA2 |
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr7_+_129906660 | 0.01 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chrX_-_49041242 | 0.01 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr12_+_123717458 | 0.01 |
ENST00000253233.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr16_+_69458537 | 0.01 |
ENST00000515314.1 ENST00000561792.1 ENST00000568237.1 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
| chrX_+_86772787 | 0.01 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr14_+_81421355 | 0.01 |
ENST00000541158.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr1_-_149982624 | 0.01 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
| chr14_+_81421861 | 0.01 |
ENST00000298171.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr2_+_48541776 | 0.01 |
ENST00000413569.1 ENST00000340553.3 |
FOXN2 |
forkhead box N2 |
| chr1_-_23520755 | 0.01 |
ENST00000314113.3 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr17_-_15244894 | 0.00 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr12_-_123717643 | 0.00 |
ENST00000541437.1 ENST00000606320.1 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
| chrY_+_16634483 | 0.00 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr14_+_81421921 | 0.00 |
ENST00000554263.1 ENST00000554435.1 |
TSHR |
thyroid stimulating hormone receptor |
| chr7_+_130020180 | 0.00 |
ENST00000481342.1 ENST00000011292.3 ENST00000604896.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr5_-_74062930 | 0.00 |
ENST00000509430.1 ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2 |
G elongation factor, mitochondrial 2 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.4 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.3 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.5 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.4 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0014718 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.8 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |