Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
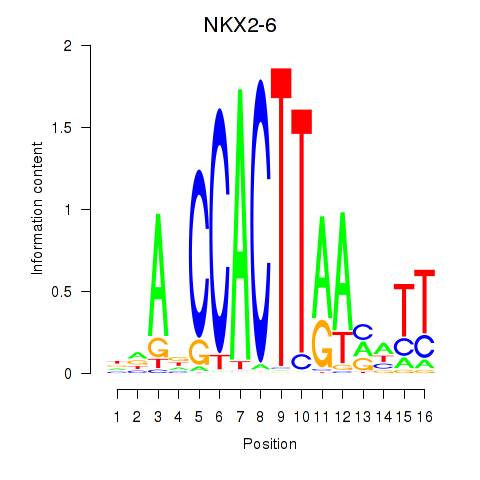
Results for NKX2-6
Z-value: 1.18
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NKX2-6 |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_121379739 | 5.43 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_15103621 | 3.35 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr15_-_55563072 | 2.27 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr6_+_33043703 | 2.16 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr11_-_76381029 | 2.05 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr7_+_142498725 | 1.99 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr22_-_37882395 | 1.91 |
ENST00000416983.3 ENST00000424765.2 ENST00000356998.3 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr10_-_6019552 | 1.63 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr10_-_6019984 | 1.56 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr1_+_158900568 | 1.50 |
ENST00000458222.1 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr22_+_23229960 | 1.46 |
ENST00000526893.1 ENST00000532223.2 ENST00000531372.1 |
IGLL5 |
immunoglobulin lambda-like polypeptide 5 |
| chr1_+_158969752 | 1.32 |
ENST00000566111.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr2_+_102928009 | 1.31 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr19_+_52076425 | 1.15 |
ENST00000436511.2 |
ZNF175 |
zinc finger protein 175 |
| chr8_-_120651020 | 1.12 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr19_+_52074502 | 1.08 |
ENST00000545217.1 ENST00000262259.2 ENST00000596504.1 |
ZNF175 |
zinc finger protein 175 |
| chr9_+_116263639 | 0.90 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_+_33914460 | 0.88 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr6_-_26216872 | 0.88 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr17_+_33914276 | 0.85 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr3_-_127541194 | 0.84 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr2_-_175462934 | 0.83 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr9_+_125133315 | 0.83 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_+_81475047 | 0.82 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr22_-_24096562 | 0.81 |
ENST00000398465.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr11_+_94277017 | 0.81 |
ENST00000358752.2 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chrX_-_47518527 | 0.80 |
ENST00000333119.3 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
| chr4_-_156875003 | 0.74 |
ENST00000433477.3 |
CTSO |
cathepsin O |
| chr9_+_125132803 | 0.71 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_41386761 | 0.67 |
ENST00000523277.2 |
GINS4 |
GINS complex subunit 4 (Sld5 homolog) |
| chrX_-_47518498 | 0.66 |
ENST00000335890.2 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
| chr9_+_134065506 | 0.66 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr5_-_111091948 | 0.65 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr4_+_17579110 | 0.62 |
ENST00000606142.1 |
LAP3 |
leucine aminopeptidase 3 |
| chr20_-_36793663 | 0.61 |
ENST00000536701.1 ENST00000536724.1 |
TGM2 |
transglutaminase 2 |
| chr13_+_115000521 | 0.61 |
ENST00000252457.5 ENST00000375308.1 |
CDC16 |
cell division cycle 16 |
| chr5_-_133510456 | 0.60 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr17_-_41116454 | 0.59 |
ENST00000427569.2 ENST00000430739.1 |
AARSD1 |
alanyl-tRNA synthetase domain containing 1 |
| chr5_-_169725231 | 0.58 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr3_+_132316081 | 0.58 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr1_+_16767167 | 0.57 |
ENST00000337132.5 |
NECAP2 |
NECAP endocytosis associated 2 |
| chr17_-_26903900 | 0.56 |
ENST00000395319.3 ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC |
aldolase C, fructose-bisphosphate |
| chr17_-_47723943 | 0.55 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chrX_+_30261847 | 0.55 |
ENST00000378981.3 ENST00000397550.1 |
MAGEB1 |
melanoma antigen family B, 1 |
| chr3_+_37035289 | 0.55 |
ENST00000455445.2 ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1 |
mutL homolog 1 |
| chr2_-_175462456 | 0.54 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr1_+_173793641 | 0.53 |
ENST00000361951.4 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr22_-_24096630 | 0.53 |
ENST00000248948.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr12_-_44152551 | 0.52 |
ENST00000416848.2 ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L |
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr20_-_1373606 | 0.51 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr10_+_121652204 | 0.50 |
ENST00000369075.3 ENST00000543134.1 |
SEC23IP |
SEC23 interacting protein |
| chr4_-_73434498 | 0.50 |
ENST00000286657.4 |
ADAMTS3 |
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr12_+_118454500 | 0.49 |
ENST00000537315.1 ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5 |
replication factor C (activator 1) 5, 36.5kDa |
| chr11_+_65627865 | 0.47 |
ENST00000308110.4 |
MUS81 |
MUS81 structure-specific endonuclease subunit |
| chr13_+_115000556 | 0.44 |
ENST00000252458.6 |
CDC16 |
cell division cycle 16 |
| chr8_+_20054878 | 0.44 |
ENST00000276390.2 ENST00000519667.1 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr1_+_16767195 | 0.43 |
ENST00000504551.2 ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2 |
NECAP endocytosis associated 2 |
| chr6_+_24775641 | 0.43 |
ENST00000378054.2 ENST00000476555.1 |
GMNN |
geminin, DNA replication inhibitor |
| chr7_+_100466433 | 0.41 |
ENST00000429658.1 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr3_-_81811312 | 0.41 |
ENST00000429644.2 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr7_-_112430647 | 0.40 |
ENST00000312814.6 |
TMEM168 |
transmembrane protein 168 |
| chr12_+_60083118 | 0.40 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_27545921 | 0.40 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr1_+_173793777 | 0.40 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr3_+_121796697 | 0.39 |
ENST00000482356.1 ENST00000393627.2 |
CD86 |
CD86 molecule |
| chr2_+_86426478 | 0.39 |
ENST00000254644.8 ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35 |
mitochondrial ribosomal protein L35 |
| chr8_-_95220775 | 0.38 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr14_+_100531615 | 0.38 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr5_-_176889381 | 0.37 |
ENST00000393563.4 ENST00000512501.1 |
DBN1 |
drebrin 1 |
| chr11_-_6640585 | 0.33 |
ENST00000533371.1 ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1 |
tripeptidyl peptidase I |
| chr3_+_37034823 | 0.33 |
ENST00000231790.2 ENST00000456676.2 |
MLH1 |
mutL homolog 1 |
| chr10_+_18629628 | 0.32 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_+_158259558 | 0.32 |
ENST00000368170.3 |
CD1C |
CD1c molecule |
| chr15_-_20170354 | 0.31 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr17_-_62493131 | 0.31 |
ENST00000539111.2 |
POLG2 |
polymerase (DNA directed), gamma 2, accessory subunit |
| chr17_+_60501228 | 0.31 |
ENST00000311506.5 |
METTL2A |
methyltransferase like 2A |
| chr3_-_46249878 | 0.31 |
ENST00000296140.3 |
CCR1 |
chemokine (C-C motif) receptor 1 |
| chr10_+_135204338 | 0.30 |
ENST00000468317.2 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
| chr15_-_72668805 | 0.29 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr9_+_131452239 | 0.29 |
ENST00000372688.4 ENST00000372686.5 |
SET |
SET nuclear oncogene |
| chr14_-_60337684 | 0.29 |
ENST00000267484.5 |
RTN1 |
reticulon 1 |
| chr3_-_114790179 | 0.29 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_+_66360665 | 0.29 |
ENST00000310190.4 |
CCS |
copper chaperone for superoxide dismutase |
| chr19_+_3178736 | 0.29 |
ENST00000246115.3 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
| chr5_+_140071178 | 0.28 |
ENST00000508522.1 ENST00000448069.2 |
HARS2 |
histidyl-tRNA synthetase 2, mitochondrial |
| chr7_+_110731062 | 0.28 |
ENST00000308478.5 ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3 |
leucine rich repeat neuronal 3 |
| chr7_-_6865826 | 0.28 |
ENST00000538180.1 |
CCZ1B |
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr9_+_131447342 | 0.28 |
ENST00000409104.3 |
SET |
SET nuclear oncogene |
| chr10_+_115939008 | 0.27 |
ENST00000369282.1 ENST00000251864.2 ENST00000369281.2 ENST00000422662.1 |
TDRD1 |
tudor domain containing 1 |
| chr19_+_7701985 | 0.26 |
ENST00000595950.1 ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2 |
syntaxin binding protein 2 |
| chr2_-_100721178 | 0.25 |
ENST00000409236.2 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr2_-_25565377 | 0.25 |
ENST00000264709.3 ENST00000406659.3 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr17_+_25621102 | 0.24 |
ENST00000581440.1 ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1 |
WD repeat and SOCS box containing 1 |
| chr19_+_17186577 | 0.24 |
ENST00000595618.1 ENST00000594824.1 |
MYO9B |
myosin IXB |
| chr1_+_110163202 | 0.23 |
ENST00000531203.1 ENST00000256578.3 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr7_+_5938386 | 0.23 |
ENST00000537980.1 |
CCZ1 |
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr12_-_71182695 | 0.22 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr8_-_121457608 | 0.22 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr14_-_106805716 | 0.21 |
ENST00000438142.2 |
IGHV4-31 |
immunoglobulin heavy variable 4-31 |
| chr11_+_98891797 | 0.21 |
ENST00000527185.1 ENST00000528682.1 ENST00000524871.1 |
CNTN5 |
contactin 5 |
| chr20_-_5100591 | 0.21 |
ENST00000379143.5 |
PCNA |
proliferating cell nuclear antigen |
| chr10_-_6019455 | 0.21 |
ENST00000530685.1 ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr16_+_69796209 | 0.20 |
ENST00000359154.2 ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_-_144416737 | 0.20 |
ENST00000367569.2 |
SF3B5 |
splicing factor 3b, subunit 5, 10kDa |
| chr12_+_44152740 | 0.20 |
ENST00000440781.2 ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4 |
interleukin-1 receptor-associated kinase 4 |
| chr7_+_5938351 | 0.20 |
ENST00000325974.6 |
CCZ1 |
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr4_+_88532028 | 0.19 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr12_-_57039739 | 0.19 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr11_-_118901559 | 0.19 |
ENST00000330775.7 ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr1_-_115259337 | 0.17 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr7_+_101917407 | 0.17 |
ENST00000487284.1 |
CUX1 |
cut-like homeobox 1 |
| chr1_-_21113105 | 0.16 |
ENST00000375000.1 ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3 |
heterochromatin protein 1, binding protein 3 |
| chr5_+_140071011 | 0.16 |
ENST00000230771.3 ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2 |
histidyl-tRNA synthetase 2, mitochondrial |
| chr17_-_7216939 | 0.15 |
ENST00000573684.1 |
GPS2 |
G protein pathway suppressor 2 |
| chr11_+_118958689 | 0.15 |
ENST00000535253.1 ENST00000392841.1 |
HMBS |
hydroxymethylbilane synthase |
| chr17_+_61562201 | 0.15 |
ENST00000290863.6 ENST00000413513.3 ENST00000421982.2 |
ACE |
angiotensin I converting enzyme |
| chr14_+_64680854 | 0.15 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr7_-_56101826 | 0.14 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chrX_-_47341928 | 0.14 |
ENST00000313116.7 |
ZNF41 |
zinc finger protein 41 |
| chr12_-_56236690 | 0.14 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr8_+_50824233 | 0.14 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr19_+_42746927 | 0.14 |
ENST00000378108.1 |
AC006486.1 |
AC006486.1 |
| chr12_+_113354341 | 0.14 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_171217677 | 0.13 |
ENST00000402921.2 |
FMO1 |
flavin containing monooxygenase 1 |
| chr14_+_50779071 | 0.12 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr15_-_56757329 | 0.12 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr8_-_121457332 | 0.12 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr15_-_65579177 | 0.12 |
ENST00000444347.2 ENST00000261888.6 |
PARP16 |
poly (ADP-ribose) polymerase family, member 16 |
| chr15_-_28344439 | 0.12 |
ENST00000431101.1 ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2 |
oculocutaneous albinism II |
| chr1_-_48866517 | 0.12 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr19_-_49527590 | 0.11 |
ENST00000357383.4 |
CGB |
chorionic gonadotropin, beta polypeptide |
| chr1_+_144989309 | 0.11 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr9_-_215744 | 0.11 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr6_-_32977345 | 0.11 |
ENST00000450833.2 ENST00000374813.1 ENST00000229829.5 |
HLA-DOA |
major histocompatibility complex, class II, DO alpha |
| chr4_-_79860506 | 0.11 |
ENST00000295462.3 ENST00000380645.4 ENST00000512733.1 |
PAQR3 |
progestin and adipoQ receptor family member III |
| chr19_-_44174330 | 0.11 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr12_-_371994 | 0.11 |
ENST00000343164.4 ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13 |
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr19_-_44174305 | 0.11 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr1_-_31902614 | 0.11 |
ENST00000596131.1 |
AC114494.1 |
HCG1787699; Uncharacterized protein |
| chr16_-_69373396 | 0.11 |
ENST00000562595.1 ENST00000562081.1 ENST00000306875.4 |
COG8 |
component of oligomeric golgi complex 8 |
| chr17_-_34890709 | 0.10 |
ENST00000544606.1 |
MYO19 |
myosin XIX |
| chr14_+_31494672 | 0.10 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr19_-_49552006 | 0.09 |
ENST00000391869.3 |
CGB1 |
chorionic gonadotropin, beta polypeptide 1 |
| chr20_-_36661826 | 0.09 |
ENST00000373448.2 ENST00000373447.3 |
TTI1 |
TELO2 interacting protein 1 |
| chr14_+_50779029 | 0.09 |
ENST00000245448.6 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr13_-_44453826 | 0.09 |
ENST00000444614.3 |
CCDC122 |
coiled-coil domain containing 122 |
| chr1_-_169337176 | 0.09 |
ENST00000472647.1 ENST00000367811.3 |
NME7 |
NME/NM23 family member 7 |
| chr7_-_150777874 | 0.09 |
ENST00000540185.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr11_-_111383064 | 0.08 |
ENST00000525791.1 ENST00000456861.2 ENST00000356018.2 |
BTG4 |
B-cell translocation gene 4 |
| chr17_-_34890732 | 0.08 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr11_-_66360548 | 0.08 |
ENST00000333861.3 |
CCDC87 |
coiled-coil domain containing 87 |
| chr9_-_70465758 | 0.08 |
ENST00000489273.1 |
CBWD5 |
COBW domain containing 5 |
| chr7_+_128116783 | 0.07 |
ENST00000262432.8 ENST00000480046.1 |
METTL2B |
methyltransferase like 2B |
| chr22_+_42017987 | 0.07 |
ENST00000405506.1 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr15_+_43425672 | 0.06 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr4_+_70916119 | 0.06 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr5_+_34757309 | 0.06 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr2_-_87088995 | 0.06 |
ENST00000393759.2 ENST00000349455.3 ENST00000331469.2 ENST00000431506.2 ENST00000393761.2 ENST00000390655.6 |
CD8B |
CD8b molecule |
| chr8_-_30706608 | 0.06 |
ENST00000256246.2 |
TEX15 |
testis expressed 15 |
| chr17_-_56606639 | 0.06 |
ENST00000579371.1 |
SEPT4 |
septin 4 |
| chr17_+_41150793 | 0.06 |
ENST00000586277.1 |
RPL27 |
ribosomal protein L27 |
| chr17_-_56606705 | 0.06 |
ENST00000317268.3 |
SEPT4 |
septin 4 |
| chr11_-_64527425 | 0.05 |
ENST00000377432.3 |
PYGM |
phosphorylase, glycogen, muscle |
| chr13_+_27998681 | 0.05 |
ENST00000381140.4 |
GTF3A |
general transcription factor IIIA |
| chr1_+_205225319 | 0.05 |
ENST00000329800.7 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
| chr14_+_21236586 | 0.05 |
ENST00000326783.3 |
EDDM3B |
epididymal protein 3B |
| chr5_+_52083730 | 0.05 |
ENST00000282588.6 ENST00000274311.2 |
ITGA1 PELO |
integrin, alpha 1 pelota homolog (Drosophila) |
| chr15_-_41120896 | 0.05 |
ENST00000299174.5 ENST00000427255.2 |
PPP1R14D |
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr17_-_56606664 | 0.04 |
ENST00000580844.1 |
SEPT4 |
septin 4 |
| chr20_-_1373682 | 0.04 |
ENST00000381724.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr16_-_20367584 | 0.04 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chr6_+_42584847 | 0.04 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_+_110162448 | 0.03 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr1_+_233086326 | 0.03 |
ENST00000366628.5 ENST00000366627.4 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
| chr5_+_140723601 | 0.03 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chrX_+_151081351 | 0.03 |
ENST00000276344.2 |
MAGEA4 |
melanoma antigen family A, 4 |
| chr11_-_61687739 | 0.03 |
ENST00000531922.1 ENST00000301773.5 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr19_+_58144529 | 0.03 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr17_+_41150479 | 0.03 |
ENST00000589913.1 |
RPL27 |
ribosomal protein L27 |
| chr1_-_175712665 | 0.03 |
ENST00000263525.2 |
TNR |
tenascin R |
| chr9_-_33402506 | 0.03 |
ENST00000377425.4 ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7 |
aquaporin 7 |
| chr21_-_31311818 | 0.02 |
ENST00000535441.1 ENST00000309434.7 ENST00000327783.4 ENST00000389124.2 ENST00000389125.3 ENST00000399913.1 |
GRIK1 |
glutamate receptor, ionotropic, kainate 1 |
| chr1_+_113009163 | 0.02 |
ENST00000256640.5 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
| chr2_-_187367356 | 0.02 |
ENST00000595956.1 |
AC018867.2 |
AC018867.2 |
| chr2_+_219246746 | 0.02 |
ENST00000233202.6 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr14_+_31494841 | 0.02 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_-_129817448 | 0.02 |
ENST00000304538.6 |
PRDM10 |
PR domain containing 10 |
| chr3_-_113956425 | 0.02 |
ENST00000482457.2 |
ZNF80 |
zinc finger protein 80 |
| chr2_+_102927962 | 0.01 |
ENST00000233954.1 ENST00000393393.3 ENST00000410040.1 |
IL1RL1 IL18R1 |
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr12_-_50290839 | 0.01 |
ENST00000552863.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr13_-_48669232 | 0.01 |
ENST00000258648.2 ENST00000378586.1 |
MED4 |
mediator complex subunit 4 |
| chr17_-_44896047 | 0.00 |
ENST00000225512.5 |
WNT3 |
wingless-type MMTV integration site family, member 3 |
| chr17_-_34890665 | 0.00 |
ENST00000586007.1 |
MYO19 |
myosin XIX |
| chr7_-_150777949 | 0.00 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr2_+_109335929 | 0.00 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.2 | 0.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 2.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 0.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 2.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 3.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 5.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 3.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.4 | 2.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 1.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.3 | 5.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 0.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 0.9 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 1.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.6 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 2.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.4 | GO:0042109 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.8 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.5 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 3.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 2.0 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 1.3 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 3.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.9 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 2.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 3.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 6.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 4.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 3.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |