Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NKX6-2
Z-value: 0.84
Transcription factors associated with NKX6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-2
|
ENSG00000148826.6 | NKX6-2 |
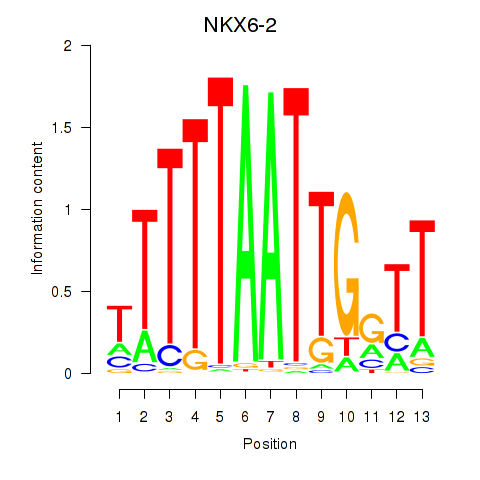
Activity profile of NKX6-2 motif
Sorted Z-values of NKX6-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_79816965 | 2.56 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr2_-_238322800 | 1.56 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr5_-_111091948 | 1.42 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr2_-_238323007 | 1.34 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr14_-_25479811 | 1.30 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr4_+_41258786 | 1.28 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr6_+_114178512 | 1.25 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr8_+_97597148 | 1.22 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr2_-_175712270 | 0.83 |
ENST00000295497.7 ENST00000444394.1 |
CHN1 |
chimerin 1 |
| chr13_-_33760216 | 0.76 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr3_-_114790179 | 0.74 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chrX_+_135230712 | 0.73 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr4_+_88896819 | 0.72 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr5_+_95066823 | 0.71 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr7_+_150065278 | 0.71 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr4_+_55095264 | 0.69 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_210501589 | 0.68 |
ENST00000413764.2 ENST00000541565.1 |
HHAT |
hedgehog acyltransferase |
| chr4_+_74269956 | 0.67 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr2_-_217560248 | 0.65 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr11_+_7618413 | 0.63 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr18_-_10701979 | 0.62 |
ENST00000538948.1 ENST00000285141.4 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chrX_+_102840408 | 0.62 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr6_-_131277510 | 0.58 |
ENST00000525193.1 ENST00000527659.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr1_+_79115503 | 0.56 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr1_-_109935819 | 0.55 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr4_+_100495864 | 0.50 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr16_+_14841923 | 0.45 |
ENST00000530328.1 ENST00000529166.1 |
NPIPA2 |
nuclear pore complex interacting protein family, member A2 |
| chr5_+_155753745 | 0.44 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr12_+_25205568 | 0.41 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr4_-_110723194 | 0.41 |
ENST00000394635.3 |
CFI |
complement factor I |
| chrX_+_105937068 | 0.40 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr9_+_82186872 | 0.40 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr10_-_63995871 | 0.39 |
ENST00000315289.2 |
RTKN2 |
rhotekin 2 |
| chr4_-_23891693 | 0.38 |
ENST00000264867.2 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr1_-_221915418 | 0.38 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr6_+_41021027 | 0.37 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr16_+_14802801 | 0.36 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr18_+_13218769 | 0.36 |
ENST00000399848.3 ENST00000361205.4 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr3_-_98241358 | 0.35 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr1_+_70820451 | 0.35 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr18_+_76829441 | 0.35 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chrX_+_68835911 | 0.35 |
ENST00000525810.1 ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA |
ectodysplasin A |
| chr2_+_211342432 | 0.34 |
ENST00000430249.2 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr3_+_178276488 | 0.33 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr16_+_31271274 | 0.32 |
ENST00000287497.8 ENST00000544665.3 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr20_+_30697298 | 0.31 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr7_-_137028498 | 0.29 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr1_+_229440129 | 0.29 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr7_-_137028534 | 0.29 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr22_+_29138013 | 0.25 |
ENST00000216027.3 ENST00000398941.2 |
HSCB |
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr8_-_57123815 | 0.25 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr1_-_21377447 | 0.25 |
ENST00000374937.3 ENST00000264211.8 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr17_+_72581057 | 0.25 |
ENST00000392620.1 |
C17orf77 |
chromosome 17 open reading frame 77 |
| chr9_+_116263778 | 0.25 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr9_-_23826298 | 0.24 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr10_-_73848086 | 0.23 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_-_22233442 | 0.23 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_174670143 | 0.22 |
ENST00000367687.1 ENST00000347255.2 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr2_+_207024306 | 0.22 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_42749759 | 0.21 |
ENST00000314073.5 |
GLTSCR1L |
GLTSCR1-like |
| chr5_+_31193847 | 0.21 |
ENST00000514738.1 ENST00000265071.2 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr1_-_150738261 | 0.21 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr1_+_174933899 | 0.20 |
ENST00000367688.3 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr3_+_52245458 | 0.20 |
ENST00000459884.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr18_+_76829385 | 0.20 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chrX_+_55744228 | 0.20 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr1_+_209878182 | 0.20 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr8_-_103876965 | 0.19 |
ENST00000337198.5 |
AZIN1 |
antizyme inhibitor 1 |
| chr9_-_110540419 | 0.19 |
ENST00000398726.3 |
AL162389.1 |
Uncharacterized protein |
| chr9_-_23825956 | 0.19 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr18_+_61575200 | 0.18 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr7_-_37024665 | 0.17 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr19_+_52772832 | 0.17 |
ENST00000593703.1 ENST00000601711.1 ENST00000599581.1 |
ZNF766 |
zinc finger protein 766 |
| chr4_+_169418255 | 0.17 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_-_110933611 | 0.17 |
ENST00000472422.2 ENST00000437429.2 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr20_-_18477862 | 0.16 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chrX_+_55744166 | 0.16 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr1_-_110933663 | 0.16 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr2_-_217559517 | 0.16 |
ENST00000449583.1 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chrX_-_48901012 | 0.16 |
ENST00000315869.7 |
TFE3 |
transcription factor binding to IGHM enhancer 3 |
| chr6_+_25963020 | 0.16 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr5_+_161275320 | 0.16 |
ENST00000437025.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr17_-_56494882 | 0.15 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr10_-_100995603 | 0.15 |
ENST00000370552.3 ENST00000370549.1 |
HPSE2 |
heparanase 2 |
| chr4_+_71337834 | 0.14 |
ENST00000304887.5 |
MUC7 |
mucin 7, secreted |
| chr14_-_60097524 | 0.14 |
ENST00000342503.4 |
RTN1 |
reticulon 1 |
| chr12_-_76462713 | 0.14 |
ENST00000552056.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr17_-_56494908 | 0.13 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr14_-_60097297 | 0.13 |
ENST00000395090.1 |
RTN1 |
reticulon 1 |
| chr22_+_24198890 | 0.13 |
ENST00000345044.6 |
SLC2A11 |
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr16_+_25228242 | 0.13 |
ENST00000219660.5 |
AQP8 |
aquaporin 8 |
| chr3_-_45838011 | 0.13 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr7_+_16828866 | 0.13 |
ENST00000597084.1 |
AC073333.1 |
Uncharacterized protein |
| chr22_-_39268308 | 0.13 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr13_+_97928395 | 0.12 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr20_+_42136308 | 0.12 |
ENST00000434666.1 ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
| chr1_-_49242553 | 0.12 |
ENST00000371833.3 |
BEND5 |
BEN domain containing 5 |
| chr17_-_7307358 | 0.11 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_-_207024134 | 0.11 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr6_+_158733692 | 0.11 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr3_+_39509163 | 0.11 |
ENST00000436143.2 ENST00000441980.2 ENST00000311042.6 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr1_+_50459990 | 0.10 |
ENST00000448346.1 |
AL645730.2 |
AL645730.2 |
| chr6_+_31582961 | 0.10 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
| chr2_+_233527443 | 0.10 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr2_-_136678123 | 0.10 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr17_-_56494713 | 0.09 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr5_-_88119580 | 0.09 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr5_+_161274940 | 0.09 |
ENST00000393943.4 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr15_+_49715293 | 0.09 |
ENST00000267843.4 ENST00000560270.1 |
FGF7 |
fibroblast growth factor 7 |
| chr2_-_207023918 | 0.09 |
ENST00000455934.2 ENST00000449699.1 ENST00000454195.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr21_-_27462351 | 0.09 |
ENST00000448850.1 |
APP |
amyloid beta (A4) precursor protein |
| chr10_-_100995540 | 0.09 |
ENST00000370546.1 ENST00000404542.1 |
HPSE2 |
heparanase 2 |
| chr1_+_115397424 | 0.09 |
ENST00000369522.3 ENST00000455987.1 |
SYCP1 |
synaptonemal complex protein 1 |
| chr7_+_138943265 | 0.09 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr7_-_102283238 | 0.09 |
ENST00000340457.8 |
UPK3BL |
uroplakin 3B-like |
| chr5_-_20575959 | 0.08 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr10_+_48189612 | 0.08 |
ENST00000453919.1 |
AGAP9 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr22_-_39268192 | 0.08 |
ENST00000216083.6 |
CBX6 |
chromobox homolog 6 |
| chr15_-_49103184 | 0.08 |
ENST00000399334.3 ENST00000325747.5 |
CEP152 |
centrosomal protein 152kDa |
| chr14_-_98444386 | 0.08 |
ENST00000556462.1 ENST00000556138.1 |
C14orf64 |
chromosome 14 open reading frame 64 |
| chr6_+_26402465 | 0.07 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr18_+_32290218 | 0.07 |
ENST00000348997.5 ENST00000588949.1 ENST00000597599.1 |
DTNA |
dystrobrevin, alpha |
| chr4_+_169418195 | 0.07 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr2_-_207024233 | 0.07 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_-_74570520 | 0.06 |
ENST00000394019.2 ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr1_+_32379174 | 0.06 |
ENST00000391369.1 |
AL136115.1 |
HCG2032337; PRO1848; Uncharacterized protein |
| chr10_-_115904361 | 0.06 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr5_+_67586465 | 0.06 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_-_226926864 | 0.06 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr4_+_169013666 | 0.05 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr7_-_102184083 | 0.05 |
ENST00000379357.5 |
POLR2J3 |
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr19_+_54641444 | 0.05 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr4_-_87374283 | 0.05 |
ENST00000361569.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr17_-_42992856 | 0.05 |
ENST00000588316.1 ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP |
glial fibrillary acidic protein |
| chr12_+_122241928 | 0.04 |
ENST00000604567.1 ENST00000542440.1 |
SETD1B |
SET domain containing 1B |
| chr5_+_161274685 | 0.04 |
ENST00000428797.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr14_+_72399833 | 0.04 |
ENST00000553530.1 ENST00000556437.1 |
RGS6 |
regulator of G-protein signaling 6 |
| chr18_-_47018769 | 0.03 |
ENST00000583637.1 ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17 |
ribosomal protein L17 |
| chr3_+_39509070 | 0.03 |
ENST00000354668.4 ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr8_+_79428539 | 0.03 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr16_+_21610879 | 0.03 |
ENST00000396014.4 |
METTL9 |
methyltransferase like 9 |
| chr10_+_26505179 | 0.02 |
ENST00000376261.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr6_-_64029879 | 0.02 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr6_-_74231444 | 0.02 |
ENST00000331523.2 ENST00000356303.2 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr16_+_31128978 | 0.02 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr15_+_79166065 | 0.01 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr2_+_218990727 | 0.01 |
ENST00000318507.2 ENST00000454148.1 |
CXCR2 |
chemokine (C-X-C motif) receptor 2 |
| chr17_-_73663245 | 0.01 |
ENST00000584999.1 ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5 |
RecQ protein-like 5 |
| chr6_-_26043885 | 0.01 |
ENST00000357905.2 |
HIST1H2BB |
histone cluster 1, H2bb |
| chr5_+_7373230 | 0.01 |
ENST00000500616.2 |
CTD-2296D1.5 |
CTD-2296D1.5 |
| chr1_+_196621002 | 0.01 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr2_+_68962014 | 0.01 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr19_+_12175504 | 0.01 |
ENST00000439326.3 |
ZNF844 |
zinc finger protein 844 |
| chr17_+_57232690 | 0.00 |
ENST00000262293.4 |
PRR11 |
proline rich 11 |
| chr12_-_53045948 | 0.00 |
ENST00000309680.3 |
KRT2 |
keratin 2 |
| chr15_-_89764929 | 0.00 |
ENST00000268125.5 |
RLBP1 |
retinaldehyde binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 1.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 1.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 0.8 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.2 | 0.7 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.2 | 0.7 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.7 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.2 | 0.6 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 1.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.3 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) response to oleic acid(GO:0034201) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0034379 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) chylomicron assembly(GO:0034378) very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 3.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.6 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.4 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |