Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
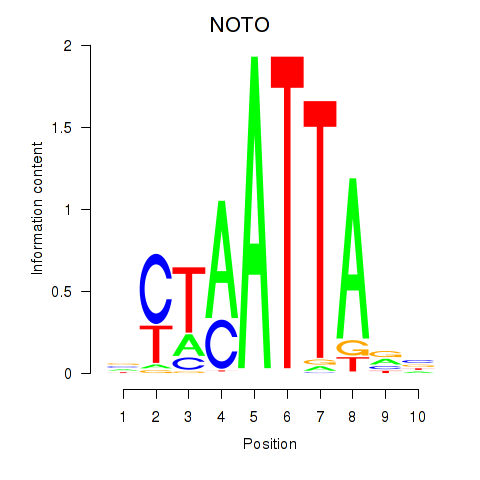
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.54
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NOTO
|
ENSG00000214513.3 | NOTO |
|
VSX2
|
ENSG00000119614.2 | VSX2 |
|
DLX2
|
ENSG00000115844.6 | DLX2 |
|
DLX6
|
ENSG00000006377.9 | DLX6 |
|
NKX6-1
|
ENSG00000163623.5 | NKX6-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX6-1 | hg19_v2_chr4_-_85419603_85419603 | -0.18 | 5.1e-01 | Click! |
| NOTO | hg19_v2_chr2_+_73429386_73429386 | -0.17 | 5.4e-01 | Click! |
| DLX2 | hg19_v2_chr2_-_172967621_172967637 | 0.15 | 5.7e-01 | Click! |
| DLX6 | hg19_v2_chr7_+_96634850_96634874, hg19_v2_chr7_+_96635277_96635301 | -0.09 | 7.5e-01 | Click! |
Activity profile of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Sorted Z-values of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NOTO_VSX2_DLX2_DLX6_NKX6-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_81110684 | 2.84 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr12_-_6233828 | 1.16 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr7_-_107642348 | 1.06 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr3_+_157154578 | 0.96 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr5_+_53751445 | 0.91 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr17_-_77924627 | 0.80 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr4_-_111563076 | 0.79 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr4_-_186696425 | 0.76 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_-_145278475 | 0.73 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_16761007 | 0.70 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr11_-_33913708 | 0.69 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_32157947 | 0.68 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr2_-_190927447 | 0.58 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr17_-_10421853 | 0.57 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr1_-_227505289 | 0.55 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr12_+_54378923 | 0.53 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr13_-_36788718 | 0.52 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr7_+_73442487 | 0.52 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr10_-_92681033 | 0.49 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr7_+_73442422 | 0.47 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr3_+_159557637 | 0.46 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr4_+_169418195 | 0.45 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_+_173116225 | 0.44 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr21_-_40033618 | 0.40 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr4_+_41614909 | 0.40 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr18_-_53089723 | 0.39 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr7_+_73442457 | 0.38 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr3_-_114477787 | 0.38 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr6_+_151646800 | 0.37 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr3_+_35722487 | 0.36 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr11_-_2160180 | 0.35 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr19_+_50016411 | 0.35 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_+_73442102 | 0.32 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chr13_-_67802549 | 0.31 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr12_+_26348582 | 0.31 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr3_-_114477962 | 0.31 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_174151536 | 0.30 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr17_-_64225508 | 0.30 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_16759711 | 0.28 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_81101277 | 0.27 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr2_-_175711133 | 0.27 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_-_27205136 | 0.27 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr6_-_36515177 | 0.27 |
ENST00000229812.7 |
STK38 |
serine/threonine kinase 38 |
| chr12_-_30887948 | 0.26 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr3_+_35721106 | 0.26 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_27169801 | 0.26 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr10_-_75415825 | 0.25 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr19_+_50016610 | 0.24 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr12_-_71031185 | 0.24 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr9_+_470288 | 0.24 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr7_-_151217166 | 0.24 |
ENST00000496004.1 |
RHEB |
Ras homolog enriched in brain |
| chr12_+_26348246 | 0.24 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr3_-_141747950 | 0.24 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_+_48243352 | 0.23 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr8_+_50824233 | 0.23 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr12_+_26348429 | 0.23 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr17_-_10325261 | 0.22 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr10_+_17270214 | 0.22 |
ENST00000544301.1 |
VIM |
vimentin |
| chr13_+_102104980 | 0.22 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr22_-_36236623 | 0.22 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr4_+_3388057 | 0.22 |
ENST00000538395.1 |
RGS12 |
regulator of G-protein signaling 12 |
| chr4_-_74486217 | 0.21 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr3_+_158787041 | 0.21 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr13_-_110438914 | 0.21 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr3_-_114790179 | 0.20 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_+_77593448 | 0.20 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr3_-_178984759 | 0.20 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_+_158991025 | 0.19 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr3_-_149293990 | 0.19 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr2_+_187454749 | 0.19 |
ENST00000261023.3 ENST00000374907.3 |
ITGAV |
integrin, alpha V |
| chr8_+_77593474 | 0.19 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr2_+_169757750 | 0.19 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr16_-_28621298 | 0.19 |
ENST00000566189.1 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chrX_-_106243451 | 0.18 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr10_+_124320156 | 0.18 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr15_+_58702742 | 0.18 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr17_-_10452929 | 0.18 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr1_+_62439037 | 0.18 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr11_-_129062093 | 0.18 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr16_+_22501658 | 0.17 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr22_-_36236265 | 0.17 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_145277569 | 0.17 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr13_-_88323218 | 0.17 |
ENST00000436290.2 ENST00000453832.2 ENST00000606590.1 |
MIR4500HG |
MIR4500 host gene (non-protein coding) |
| chr10_+_124320195 | 0.17 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr4_+_71457970 | 0.17 |
ENST00000322937.6 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr13_+_102104952 | 0.17 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr18_-_53069419 | 0.16 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr2_-_145275228 | 0.16 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_89746173 | 0.16 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr12_-_51422017 | 0.16 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chrX_+_135251783 | 0.16 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr7_-_143599207 | 0.15 |
ENST00000355951.2 ENST00000479870.1 ENST00000478172.1 |
FAM115A |
family with sequence similarity 115, member A |
| chr21_-_35899113 | 0.15 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr16_-_29910853 | 0.15 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr16_-_28621312 | 0.15 |
ENST00000314752.7 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr6_-_49604545 | 0.15 |
ENST00000371175.4 ENST00000229810.7 |
RHAG |
Rh-associated glycoprotein |
| chr4_+_86396265 | 0.15 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr6_-_100912785 | 0.15 |
ENST00000369208.3 |
SIM1 |
single-minded family bHLH transcription factor 1 |
| chr20_-_56285595 | 0.15 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr12_-_71031220 | 0.15 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr16_-_28621353 | 0.15 |
ENST00000567512.1 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr7_+_138145076 | 0.14 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr9_+_136501478 | 0.13 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr16_+_69345243 | 0.13 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr2_-_152589670 | 0.13 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr2_+_109204743 | 0.13 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_-_91296602 | 0.13 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr7_-_151217001 | 0.13 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr11_+_71900703 | 0.13 |
ENST00000393681.2 |
FOLR1 |
folate receptor 1 (adult) |
| chr9_-_13165457 | 0.13 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr14_-_61116168 | 0.13 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chrX_+_135251835 | 0.13 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr11_+_71900572 | 0.13 |
ENST00000312293.4 |
FOLR1 |
folate receptor 1 (adult) |
| chr4_-_74486347 | 0.13 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_-_91296737 | 0.13 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr15_+_91411810 | 0.13 |
ENST00000268171.3 |
FURIN |
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_36621529 | 0.12 |
ENST00000316156.4 |
MAP7D1 |
MAP7 domain containing 1 |
| chrX_+_135252050 | 0.12 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr12_+_21207503 | 0.12 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_+_206557366 | 0.12 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_-_157824292 | 0.11 |
ENST00000483851.2 |
SHOX2 |
short stature homeobox 2 |
| chr9_-_136933134 | 0.11 |
ENST00000303407.7 |
BRD3 |
bromodomain containing 3 |
| chr6_-_161695042 | 0.11 |
ENST00000366908.5 ENST00000366911.5 ENST00000366905.3 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr21_-_39870339 | 0.11 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr7_-_99717463 | 0.11 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr18_-_53257027 | 0.11 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr6_-_31107127 | 0.11 |
ENST00000259845.4 |
PSORS1C2 |
psoriasis susceptibility 1 candidate 2 |
| chrX_-_20236970 | 0.11 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr1_-_94147385 | 0.10 |
ENST00000260502.6 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr18_-_53177984 | 0.10 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr14_-_78083112 | 0.10 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr12_+_49621658 | 0.10 |
ENST00000541364.1 |
TUBA1C |
tubulin, alpha 1c |
| chr1_-_68698222 | 0.10 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_-_101360331 | 0.10 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr12_-_56694142 | 0.10 |
ENST00000550655.1 ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS |
citrate synthase |
| chr19_+_13049413 | 0.09 |
ENST00000316448.5 ENST00000588454.1 |
CALR |
calreticulin |
| chr14_-_101295407 | 0.09 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chrX_-_64196351 | 0.09 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr11_+_125034586 | 0.09 |
ENST00000298282.9 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
| chr15_-_31393910 | 0.09 |
ENST00000397795.2 ENST00000256552.6 ENST00000559179.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_-_99716952 | 0.09 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr9_-_14314518 | 0.09 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr9_-_14314566 | 0.09 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr14_+_22977587 | 0.09 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr14_+_61654271 | 0.09 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr17_+_48823975 | 0.09 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr16_-_66764119 | 0.09 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr3_-_58613323 | 0.08 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr5_+_66300446 | 0.08 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_201173667 | 0.08 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr1_-_94079648 | 0.08 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr11_-_13517565 | 0.08 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr4_+_41614720 | 0.08 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr15_+_89631381 | 0.08 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr8_+_26150628 | 0.08 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr6_-_161695074 | 0.08 |
ENST00000457520.2 ENST00000366906.5 ENST00000320285.4 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_-_55237484 | 0.08 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr2_+_171034646 | 0.08 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr1_-_94586651 | 0.08 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr13_+_53602894 | 0.08 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr1_-_95391315 | 0.07 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr1_-_190446759 | 0.07 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr15_+_58430567 | 0.07 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr9_+_77230499 | 0.07 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr5_-_141249154 | 0.07 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chrX_-_64196376 | 0.07 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr13_+_109248500 | 0.07 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr10_-_33623310 | 0.07 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr6_-_33160231 | 0.07 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chr6_+_151042224 | 0.07 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_-_99279928 | 0.07 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr14_-_37051798 | 0.07 |
ENST00000258829.5 |
NKX2-8 |
NK2 homeobox 8 |
| chr7_+_107224364 | 0.07 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chrX_+_22050546 | 0.07 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr10_-_116444371 | 0.07 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr5_-_88119580 | 0.07 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr6_+_39760129 | 0.07 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr3_-_33686743 | 0.07 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr1_-_204329013 | 0.06 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr2_-_88285309 | 0.06 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr12_+_20963632 | 0.06 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chrX_-_64196307 | 0.06 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr8_-_133637624 | 0.06 |
ENST00000522789.1 |
LRRC6 |
leucine rich repeat containing 6 |
| chr7_-_87342564 | 0.06 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr9_-_124989804 | 0.06 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr11_+_65554493 | 0.06 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr8_+_24298531 | 0.06 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr3_+_46919235 | 0.06 |
ENST00000449590.1 |
PTH1R |
parathyroid hormone 1 receptor |
| chr12_-_118796910 | 0.06 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr8_-_72268889 | 0.06 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr20_+_60174827 | 0.06 |
ENST00000543233.1 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
| chr8_-_17579726 | 0.06 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr1_+_214161272 | 0.06 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr12_+_20963647 | 0.06 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr5_-_58295712 | 0.06 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr5_+_176811431 | 0.06 |
ENST00000512593.1 ENST00000324417.5 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr15_+_96869165 | 0.05 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr5_-_134871639 | 0.05 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chrX_-_15288154 | 0.05 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chrX_+_46771711 | 0.05 |
ENST00000424392.1 ENST00000397189.1 |
PHF16 |
jade family PHD finger 3 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.3 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 1.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 3.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 3.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 1.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.3 | 1.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 0.8 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 1.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.5 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.3 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0090611 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) glycerol transport(GO:0015793) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0016199 | axon choice point recognition(GO:0016198) axon midline choice point recognition(GO:0016199) |
| 0.0 | 1.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.0 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |